Abstract
The complete mitochondrial DNA (mtDNA) sequences of two endemic subspecies of the White spotted charr (Salvelinus leucomaenis) in Japan were determined. The complete mtDNA sequences of two individuals of S. l. imbrius and S. l. pluvicus were analyzed and compared with those of other charrs in GenBank. The whole mtDNA sequences of S. l. imbrius and S. l. pluvicus were 16,655 bp in length. The whole mtDNA sequence comparisons between S. leucomaenis in Japan and other charrs in GenBank, including charrs from East Asia to North American, revealed that S. l. imbrius and S. l. pluvius in Japan belonged to a different group from S. curilus (syn. S. alma krascheninnikovi), which is sympatric with S. leucomaenis in Hokkaido. Furthermore, it was discovered that the Japanese subspecies had 8 nucleotide deletions and four nucleotide insertions compared with S. curilus.
The charr is an anadromous freshwater fish in the salmonid family that is widely distributed in the Northern Hemisphere. In Japan, the white spotted charr (Salvelinus leucomaenis) is distributed in Hokkaido Island and Honshu Island (Kawanabe Citation1989). Salvelinus leucomaenis in Japan has been classified into four subspecies (S. l. imbrius, S. l. japonicus, S. l. leucomaenis, and S. l. pluvicus) based on their zoogeographic patterns and morphological characteristics (Hosoya Citation2013). The complete mitochondrial DNA (mtDNA) sequences of endemic subspecies of S. leucomaenis in Japan have not been established and analyzed in detail. In this study, to assess the phylogeographic patterns of these species, we examined the complete mtDNA sequences of S. l. imbrius and S. l. pluvicus and compared them with those of other charrs from East Asia to North American.
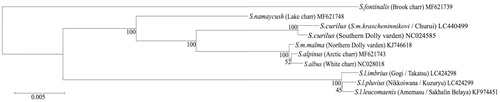
The S. l. imbrius specimen was collected in the upstream of Takatsu river (34.696620N, 131.820622E) and the S. l. pluvicus was collected in the upstream of Kuzuryu river (36.219903N,136.131935E) by fly fishing and stored in a frozen state (in Seikei Junior and Senior High School). The mtDNA was extracted, and five primer sets for its amplification. After long PCR with HiFi DNA polymerase (Greiner Bio-One, Frickenhausen, Germany), the PCR products were sequenced using a BigDye Terminator Cycle Sequencing Ready Reaction Kit (Applied Biosystems, CA, USA) and an ABI PRISM 3100 Genetic Analyzer (Applied Biosystems). The whole mtDNA sequences of S. l. imbrius and S. l. pluvicus were 16,655 bp in length and submitted to GenBank (Access No. LC424298 for S. l. imbrius, and LC424299 for S. l. pluvicus). For use in phylogenetic analyses, the mtDNA sequence of S. curilus from Churui river (43.727766N,145.091254E) was also determined and submitted to GenBank (Access No. LC440499). S. curilus (syn. S. alma krascheninnikovi) has a sympatric habitat with S. leucomaenis in Hokkaido. Comparison of the mtDNA sequences between S. l. imbrius, S. l. pluvicus, and S. curilus revealed that the Japanese subspecies had 8 nucleotide deletions and four nucleotide insertions in the CR, 16S rRNA, and OriL. Multiple mtDNA sequences comparisons were performed to evaluate the levels of similarity and difference between nucleotides using MEGA software version 7. A phylogenetic evaluation was conducted by the maximum likelihood (ML) method with 1000 bootstrap simulations using the same MEGA software (Hall Citation2013). The ML tree showed that the White spotted charrs (S. l. imbrius and S. l. pluvius) in Japan belonged to a different group from S. curilus (). This probably arose by differences in the species of direct origin and their migration routes. Furthermore, S. l. imbrius and S. l. pluvius were in the same cluster as S. l. leucomaenis in Sakhalin Belaya (KF 974451), with S. l. imbrius branching from the cluster at the earliest time. In addition, the cluster containing S. curilus (syn. S. malma Krascheninikovi) and the cluster containing S. alpinus were different. Notably, the three kinds of braches in the latter cluster had a similar appearance to that of a subspecies within the cluster containing S. leucomaenis. Among the target species in this study, S. fontinalis branched at the earliest time, and the closest species to S. leucomaenis was S. namaycush in North America.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Hall BG. 2013. Building phylogenetic trees from molecular data with MEGA. Mol Biol Evol. 30:1229–1235.
- Hosoya K. 2013. Fishes of Japan with pictorial keys to the species. 3rd ed. Tokyo: Tokai University Press (in Japanese); p. 362–367, 1833.
- Kawanabe H. 1989. Japanese char(r(r))s and masu-salmon problems: a review. Physiol Ecol Jpn Spec. 1:13–24.