Abstract
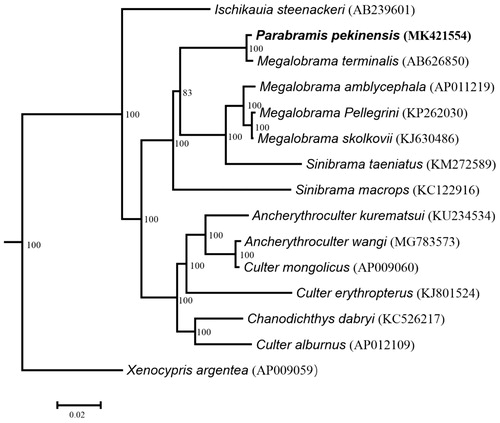
The complete mitochondrial genome of Parabramis pekinensis was determined in this study. It contained 13 protein-coding genes (PCGs), 22 tRNA, two rRNAs, and a control region with the base composition 31.11% A, 27.80% C, 24.81% T, and 16.27% G. Here, we compared this newly determined mitogenome with another one from the same species reported before. The variable sites and the genetic distances between the two mitogenomes were 96 bp and 0.6%. Eight-three variable sites were occurred in the PCGs. The results from the phylogenetic analysis showed that P. pekinensis is close to Megalobrama terminalis.
Parabramis is a monotypic genus within subfamily Culterinae in Cyprinidae. Parabramis pekinensis (Teleostei, Cypriniformes), known as ‘white Amur bream’, is native to East Asia (Froese and Pauly Citation2018). It is an important food source in this region. Here, the complete mitochondrial genome sequence of P. pekinensis was determined (GenBank accession No. MK421554) and was compared with another P. pekinensis mitogenome data reported before (Zhang et al. Citation2014). The specimens were collected from Gaoyang Town (31.098108 N, 108.675528 E), Yunyang County, Chongqing, China, and were stored in the museum of the Key Laboratory of Freshwater Fish Reproduction and Development (Southwest University, China). Primers used in this study and methods for collecting DNA sequences followed the procedures outlined in Wang et al. (Citation2011).
The complete mitochondrial genome of P. pekinensis was a circular molecule with 16,623 bp in length. It contained 13 PCGs, 22 tRNA genes, two rRNA genes, and one control region. Most of the genes were encoded on H-strand, whereas ND6 and eight tRNA genes were encoded on L-strand. The overall nucleotide composition was 31.11% A, 24.81% T, 27.80% C, and 16.27% G, with a slight AT bias. All the mitochondrial PCGs in the P. pekinensis use the standard ATG start codon, except for CO1, which utilizes GTG. Seven PCGs contain TAA stop codon and four contain TAG stop codon, whereas two contain the incomplete stop codon T– –.
Comparing with another mitogenome of P. pekinensis, the length of it was 16,622 bp (accession No. JX242531; Zhang et al. Citation2014). The genetic distance between the two mitogenomes was 0.6%. One indel and 96 nucleotide substitutions were found between the two mitogenomes. Among these substitutions, 88 transitions (91.7%) and eight transversions (8.3%) were found. There were 83 variable sites appeared in the PCGs and five sites in control region. The other variable sites were occurred in 12S rRNA, 16S rRNA, and tRNAs.
To confirm the phylogenetic position of P. pekinensis among Culterinae, phylogenetic analysis was performed based on the complete mtDNA using maximum-likelihood and Bayesian methods. The results from the analyses show that P. pekinensis demonstrate a close relationship with Megalobrama terminalis.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Froese R, Pauly D, editors. 2018. FishBase. World Wide Web electronic publication. Version (06/2018).
- Wang J, Li P, Zhang Y, Peng Z. 2011. The complete mitochondrial genome of Chinese rare minnow, Gobiocypris rarus (Teleostei: Cypriniformes). Mitochondrial DNA. 22:178–180.
- Zhang X, Song W, Wang Y, Du R, Wang W. 2014. The complete nucleotide sequence of white Amur bream (Parabramis pekinensis) mitochondrial genome. Mitochondrial DNA. 25:361–362.