Abstract
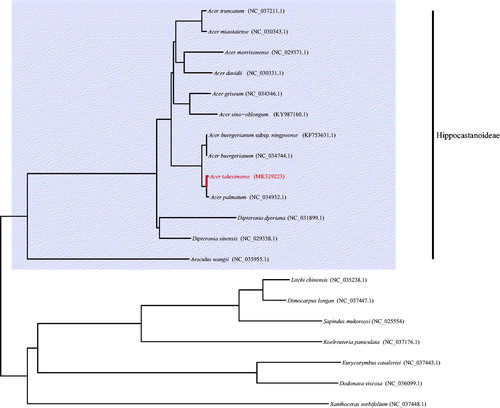
The complete chloroplast (cp) genome sequence of Acer takesimense, a member of maple tree and an endemic species to Ulleung Island of Korea, was firstly analyzed in this study. It was formed a typical circular structure of 157,023 bp in length and composed of a large single copy region (85,371 bp) and a small single copy region (18,160 bp) which were separated by two inverted repeat regions (26,746 bp). Interestingly, rps2 gene was pseudogene due to two deletions of 5 bp. In the phylogenetic tree using chloroplast genome sequence, it was clear that the genus Acer, a monophyletic group, was sister to Dipteronia in the subfamily Hippocastanoideae and A. takesimense was a sister group of A. palmatum.
The genus Acer L., well-known as maple trees and the largest tree genus in the Northern Hemisphere besides oak tree (Quercus), is widely distributed from Europe to North America with the center of species diversity in the central and eastern Asia. It was classified to the family Aceraceae with Dipteronia but now it is recognized as a member of the family Sapindaceae in APG IV (2016) system. It includes over 120 species and has been suggested different infra-generic classification systems composed of 12-25 sections which are still unclear the relationships among them (Suh et al. Citation2000; Grimm et al. Citation2006; Li et al. Citation2006). Acer takesimense is an endemic to Ulleng Island, where located at the eastern sea of Korea and is famous place of its high endemism. For understanding the genomic feature of this plant, we analyzed the complete chloroplast genome sequence.
We collected the plant material from Bongrae-dong of Ulleung Island and the voucher was deposited at the Herbarium of Kyungpook National University (KNU). Complete chloroplast genome of Acer takesimense (MK329223) was sequenced by HiSeq4000 of Illumina. Totally 51,682,020 paired-end reads (2x151bp) were obtained and 226,486 reads were assembled to the reference chloroplast genomes after reads end trimming with an error probability limit of 0.01. And then assembled reads were de novo assembled using the Geneious assembler. Using the assembled contigs, we conducted to align and repeat the procedure up to make a single contig. Complete chloroplast genome was annotated using Geneious 10.2.6 (Kearse et al. Citation2012) with manual correction and tRNAScan-SE (Lowe and Eddy Citation1997) for tRNA gene. The average coverage of this chloroplast genome was 210.2. The phylogenetic tree was constructed with related Asteraceae members based on the concatenated 78 coding genes using RAxML (Stamatakis Citation2014)
It was typical circular form with 157,023 bp in length and comprised a large single copy region (LSC, 85,371 bp), a small single copy region (SSC, 18,160 bp), and two inverted repeat regions (IR, 26,746 bp). It was composed of 134 genes and they were identified 86 coding genes, eight rRNA genes, 37 tRNA genes, and three pseudogenes. rps2 gene was pseudogene as well as ycf1 and trnH, which were positioned in the boundary area of IR because of two deletions of 5 bp. In the phylogenetic tree conducted using the protein coding genes, the monophyly of the genus was strongly supported and A. takesimense was a sister group of A. palmatum (KY457568). And also, it was clear the sister relationship with the genus Dipteronia in the subfamily Hippocastanoideae (). Comparing to A. palmatum chloroplast genome, it was 344bp shorter in length, especially in the inter genic spacer regions of LSC. From the result, it is expected to provide useful information for understanding the diversification and developing the molecular marker in the genus Acer.
Disclosure statement
The authors have no conflicts of interest about this study.
Additional information
Funding
References
- Grimm GW, Renner SS, Stamatakis A, Hemleben V. 2006. A nuclear ribosomal DNA phylogeny of Acer inferred with maximum likelihood, splits graphs, and motif analysis of 606 sequences. Evol Bioinform Online. 2:7–22.
- Kearse M, Moir R, Wilson A, Stone-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Li J, Yue J, Shoup S. 2006. Phylogenetics of Acer (Aceroideae, Sapindaceae) based on nucleotide sequences of two chloroplast non-coding regions. Harv Pap Bot. 11:101–115.
- Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955–964.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Suh Y, Heo K, Park CW. 2000. Phylogenetic relationships of Maples (Acer L.; Aceraceae) implied by nuclear ribosomal ITS sequences. J Plant Res. 113:193–202.