Abstract
Hemibagrus spilopterus is a tropical freshwater fish with high nutritional value. Here, we present the complete mitogenome of H. spilopterus. It was a circular molecule with 16,521 bp in length and contained 22 tRNA genes, 13 protein-coding genes, 2 rRNA genes, and a control region. The overall base composition was 31.68% A, 26.55% T, 26.77% C, and 15.01% G, with an AT content of 58.23%. The phylogenetic analysis showed that it was closely related to Hemibagrus sp. RCH and the genus Hemibagrus was not a monophyletic group.
The genus Hemibagrus (Teleostei: Siluriformes: Bagridae) consists of more than 40 valid species (Froese and Pauly Citation2019), presently, however, only six complete mitochondrial genome (mitogenome) data were available. Here, we determined the complete mitogenome sequence of Hemibagrus spilopterus, a tropical freshwater bagrid catfish with high-nutritional value and distributed in the lower Mekong River basin in Cambodia (Ng and Rainboth Citation1999). Samples were collected from Siem Reap in Cambodia (13.240391 N, 103.823090 E) and the specimens were deposited in the Southwest University Museum of Freshwater Fishes. The same approach as described in Zeng et al. (Citation2012) was used to obtain the complete mitogenome of H. spilopterus. The sequences were aligned and analyzed using MEGA7 (Kumar et al. Citation2016). The phylogenetic analyses based on the maximum likelihood (ML) method and Bayesian inference (BI) were conducted using PhyML 3.0 (Guindon et al. Citation2010) and MrBayes 3.2.6 (Ronquist et al. Citation2012), respectively.
The complete mitogenome sequence of H. spilopterus was a circular molecule with 16,521 bp in length (GenBank accession No. JQ343983). It contained 22 tRNA genes, 13 protein-coding genes (PCGs), 2 ribosomal RNA genes, and a control region, which was identical to the other typical vertebrates (Peng et al. Citation2006). The overall base composition was 31.68% A, 26.55% T, 26.77% C, and 15.01% G, with an AT content of 58.23%. Some overlapping sites existed in PCGs, including the four notable overlaps between ATP8 and ATP6 (by 10 bp), ATP6 and COIII (by 1 bp), ND4L and ND4 (by 7 bp), and ND5 and ND6 (by 4 bp). Among the 13 PCGs, all genes used ATG as the start codon except COI, which used GTG. Six PCGs (ND1, COI, ATP8, ATP6, ND4L, and ND5) used TAA as stop codon; three PCGs (ND2, ND3, and ND6) used TAG as stop codon; while four PCGs (COII, COIII, ND4, and Cyt b) had incomplete stop codon T–. The length of the tRNAs ranged from 67 bp (tRNA-Cys and tRNA-Ser) to 75 bp (tRNA-Leu), which was consistent with the mitogenome of other species within the genus Hemibagrus (Zeng et al. Citation2012).
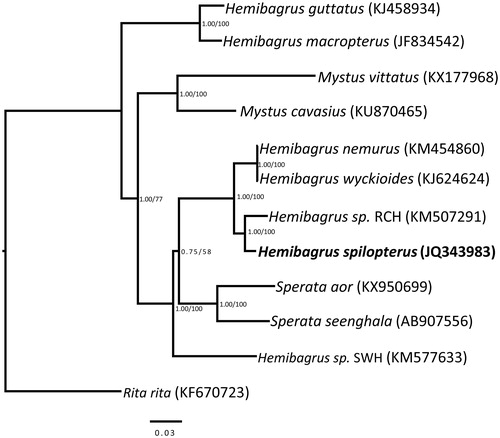
The results from the phylogenetic analysis strongly supported that the genus Hemibagrus is not a monophyletic group and H. spilopterus had a close relationship with Hemibagrus sp. RCH (). In conclusion, the complete mitogenome of H. spilopterus can provide essential nucleotide data for further evolutionary analysis in family Bagridae.
Acknowledgments
We thank Ms. Dalin Ly (Royal University of Agriculture, Cambodia) for help in field sampling. We also thank Mr. Qing Zeng for the laboratory work.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Froese R, Pauly D, editors. 2019. FishBase. World Wide Web electronic publication. version (02/2019). http://www.fishbase.org.
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 59:307–321.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- Ng HH, Rainboth WJ. 1999. The bagrid catfish genus Hemibagrus (Teleostei: Siluriformes) in central Indochina with a new species from the Mekong River. Raffles Bull Zool. 47:555–576.
- Peng Z, Wang J, He S. 2006. The complete mitochondrial genome of the helmet catfish Cranoglanis bouderius (Siluriformes: Cranoglanididae) and the phylogeny of otophysan fishes. Gene. 376:290–297.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61:539–542.
- Zeng Q, Ye H, Peng Z, Wang Z. 2012. Mitochondrial genome of Hemibagrus macropterus (Teleostei, Siluriformes). Mitochondrial DNA. 23:355–357.