Abstract
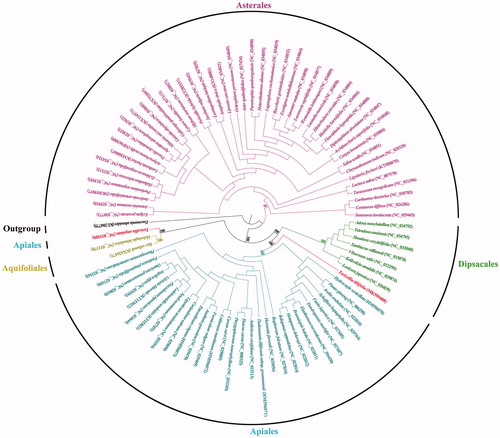
In this study, we presented the first complete plastome of Toricellia tiliifolia, an Arcto-Tertiary relict endemic to eastern Asian, using Illumina pair-end sequencing data. The plastome was 158,627 bp in length and consisted of a pair of inverted repeats (IRs 26,174 bp), which were separated by a large single-copy region (LSC 87,847 bp) and a small single-copy region (SSC 18,432 bp). The plastome harbored 114 unique genes, including 80 protein-coding genes, 30 tRNAs, and 4 rRNAs. Phylogenetic analysis indicated that T. tiliifolia was the earliest diverged clade within the order Apiales.
Toricellia DC., belonging to the family Toricelliaceae, is represented by only two living species, namely, Toricellia angulata Oliv. and T. tiliifolia DC. (Xiang and David Citation2005). The genus is now endemic to eastern Asia, while it has extensive fossil records throughout the Northern Hemisphere during the Paleocene and Eocene (Manchester et al. Citation2009). The contractive distribution of the genus may have been triggered by the Neogene climate cooling and the Pleistocene glaciations (Qian and Ricklefs Citation2000; Manchester et al. Citation2009). The genus thus is an excellent example of Arcto-Tertiary relicts to give insights into the evolution of the Arcto-Tertiary flora in the Northern Hemisphere. To date, the complete plastome of T. angulata has been sequenced by Jia and Zhang (Citation2017). Here, we report the whole plastome of another species, T. tiliifolia.
Healthy and fresh leaves of T. tiliifolia were collected from Gengma county (23°32′14′′N, 99°23′34′′E), Yunnan province, China. Voucher specimen (LCK-20190113) was deposited in the Herbarium of Kunming Institute of Botany, Chinese Academy of Sciences (KUN). Total genomic DNA was extracted from 100 mg of fresh leaves using a modified CTAB method (Yang et al. Citation2014). Subsequently, we amplified platome DNA regions using the method developed by Yang et al. (Citation2014). Purified DNA was used to construct libraries according to the manufacturer’s manual (Illumina, San Diego, California, USA). Ensuing, the libraries were sequenced using the Illumina Hiseq 2000 sequencing platform at BGI (Shenzhen, Guangdong, China). We assembled the plastome following the method described by Jin et al. (Citation2018). Plastome of T. angulata (Genbank accession: KX648359) was used as reference sequence. The assembled genome was annotated in Geneious 7.0 (Kearse et al. Citation2012), and manually checked for start and stop codons and intron/exon boundaries. The validated complete plastome of T. tiliifolia deposited in GenBank under accession number MK599408.
The T. tiliifolia plastome was 158,627 bp in length and presented a typical quadripartite structure consisting of one large single-copy region (LSC, 87,847 bp), one small single-copy region (SSC, 18,432 bp), and a pair of inverted repeat regions (IRs, 26,174 bp). The plastome encoded 134 genes, of which 114 were unique genes (80 protein-coding genes, 30 tRNAs, and 4 rRNAs). Among these unique genes, 12 protein-coding genes (atpF, ndhA, ndhB, petB, petD, rpl16, rpl2, rpoC1, rps12, rps16, clpP, and ycf3) and 6 tRNAs (trnA-UGC, trnG-UCC, trnI-GAU, trnK-UUU, trnL-UAA, and trnV-UAC) contained at least one intron.
The phylogenetic analysis was performed based on 87 completed campanulids plastomes using the software RAxML version 8.2.11 with 1000 bootstrap replicates (Stamatakis Citation2014). The phylogeny robustly supported that T. tiliifolia was the earliest diverged clade within the order Apiales. Interestingly, T. tiliifolia did not form a monophyletic group with its congener, T. angulata ().
Disclosure statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Additional information
Funding
References
- Jia XM, Zhang HL. 2017. Characterization of the complete chloroplast genome of the Chinese endemic plant Toricellia antulata (Apiales: Torricelliaceae). Conservation Genet Resour. 9:221–224.
- Jin JJ, Yu WB, Yang JB, Song Y, Yi TS, Li DZ. 2018. GetOrganelle: a simple and fast pipeline for de novo assembly of a complete circular chloroplast genome using genome skimming data. bioRxiv. doi: 10.1101/256479.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Manchester SR, Chen ZD, An-Ming LU, Uemura K. 2009. Eastern Asian endemic seed plant genera and their paleogeographic history throughout the northern hemisphere. J Syst Evol. 47:1–42.
- Qian H, Ricklefs RE. 2000. Large-scale processes and the Asian bias in species diversity of temperate plants. Nature. 407:180–182.
- Stamatakis A. 2014. Raxml version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Xiang QY, David EB. 2005. Toricelliaceae. In: Wu ZY, Raven PH, editors. Flora of China, Vol. 14. Beijing and St. Louis: Science Press and Missouri Botanical Garden Press; p. 233–234.
- Yang JB, Li DZ, Li HT. 2014. Highly effective sequencing whole chloroplast genomes of angiosperms by nine novel universal primer pairs. Mol Ecol Notes. 14:1024–1031.