Abstract
The Korean crevice salamander, Karsenia koreana, is the only known plethodontid salamander in Asia. In this study, we provide the first report of this salamander’s complete mitochondrial genome. The mitogenome had a length of 16,780 bp and consisted of 13 protein-coding genes, 2 rRNA genes, 22 tRNA genes, and 2 noncoding regions. The presented sequence may be highly useful for future phylogenetic and evolutionary studies of Plethodontidae.
The Korean crevice salamander (Karsenia koreana) is the only known Asian plethodontid salamander and is mainly distributed in central South Korea (Min et al. Citation2005; Lee and Park Citation2016). Despite multiple recent phylogenetic studies (Vieites et al. Citation2011; Shen et al. Citation2016; Wake Citation2017), this salamander’s phylogenetic location within Plethodontidae and how the current biogeographic distribution of this species was established have not been thoroughly elucidated to date. In 2011, Vieites et al. reported a partial mitochondrial genome sequence of K. koreana; this sequence did not include some cytochrome b sequences, the complete sequences of two tRNAs (tRNA-Pro and -Thr), and a displacement loop region (Vieites et al. Citation2011). In this study, we provide the first report of the complete mitochondrial genome of K. koreana, which may be useful for future phylogenetic and evolutionary studies of Plethodontidae.
We obtained the complete mitogenome from a specimen of K. koreana collected on Woraksan Mountain in Danyang, Chungbuk, South Korea (36.90°N and 128.28°E). The specimen was preserved in 70% ethanol and stored at the herpetology laboratory of Kangwon National University (Voucher No. G391KK). We extracted whole genomic DNA from the tail tissue of the specimen using Qiagen’s DNeasy Blood & Tissue kit (Qiagen Korea Ltd., Seoul, South Korea) following the manufacturer’s instructions. The whole mitochondrial DNA sequence was sequenced using the HiSeq 2000 platform (Illumina, San Diego, USA). To assemble and annotate the full mitochondrial DNA sequence, we used Geneious 9.0.4 (Biomatters Ltd., Auckland, New Zealand), tRNA Scan-SE1.21 software (http://lowelab.ucsc.edu/tRNA Scan-SE/) (Lowe and Eddy Citation1997), and Dual Organellar GenoMe Annotator (Wyman et al. Citation2004). The phylogenetic tree of K. koreana and related plethodontid salamanders was constructed with PAUP* v4.0b10 (Swofford Citation2003) using the complete mitogenome sequence obtained in this study and complete mitogenome sequences from GenBank (). We applied Bayesian inference and maximum likelihood methods to construct the tree using 1000 bootstrap replicates.
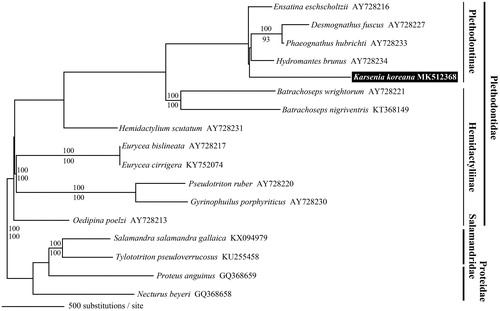
Figure 1. Maximum likelihood tree based on the complete mitochondrial genomes of Karsenia koreana and 12 other plethodontid salamanders. We used two species from each of Salamandridae and Proteidae as outgroup taxa. The accession number of each mitogenome is presented after the scientific name of the species. On each branch, the Bayesian posterior probability (above) and bootstrap value (below) are presented.
The mitogenome length of K. koreana was 16,780 bp (GenBank Accession No. MK512368). The base composition of the mitogenome was 33.3% A, 30.6% T, 13.3% G, 22.8% C, revealing an A-T rich (63.9%) structure. The mitogenome contained 13 protein-coding genes, 2 rRNA genes (12S and 16S rRNA), 22 tRNA genes, and 2 noncoding regions of an L-strand replication origin and a displacement loop region. The arrangement pattern and transcription directions of the mitogenome were identical to those of other plethodontid salamanders (Hydromantes brunus and Desmognathus wrighti) reported previously (Mueller et al. Citation2004).
The phylogenetic tree of K. koreana and related plethodontid salamanders was consistent with the finding of Shen et al. (Citation2016) that K. koreana and the genus Hydromantes in Plethodontinae are not sister taxa. In contrast, Vieites et al. (Citation2011) identified K. koreana and Hydromantes as sister taxa, possibly due in part to their inclusion of only partial tRNA genes and lack of noncoding regions in their analysis. In the present study, the high Bayesian posterior probabilities and bootstrap values of the phylogenetic tree provide strong support for the inclusion of K. koreana in Plethodontinae ().
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Lee JH, Park DS. 2016. The encyclopedia of Korean amphibians. Seoul (KR): Nature and Ecology.
- Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955–964.
- Min MS, Yang SY, Bonett RM, Vieites DR, Brandon RA, Wake DB. 2005. Discovery of the first Asian plethodontid salamander. Nature. 435;87–90.
- Mueller RL, Macey JR, Jaekel M, Wake DB, Boore JL. 2004. Morphological homoplasy, life history evolution, and historical biogeography of plethodontid salamanders inferred from complete mitochondrial genomes. PNAS. 101:13820–13825.
- Shen XX, Liang D, Chen MY, Mao RL, Wake DB, Zhang P. 2016. Enlarged multilocus data set provides surprisingly younger time of origin for the Plethodontidae, the largest family of salamanders. Syst Biol. 65:66–81.
- Swofford DL. 2003. PAUP*: Phylogenetic Analysis Using Parsimony, Version 4.0 b10. https://paup.phylosolutions.com/
- Vieites DR, Román SN, Wake MH, Wake DB. 2011. A multigenic perspective on phylogenetic relationships in the largest family of salamanders, the Plethodontidae. Mol Phylogenet Evol. 59:623–635.
- Wake DB. 2017. Persistent plethodontid themes: species, phylogenies, and biogeography. Herpetologica. 73:242–251.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
