Abstract
Halenia corniculata, characterized by having yellow corollas with spurs at the base within Halenia, is an endangered species in Korea, which represents the southern limit of the distribution range of the species. We present the complete chloroplast genome of H. corniculata. Its length is 153,198 bp and it has four subregions; 83,252 bp of large-single-copy and 18,372 bp of small-single-copy regions are separated by 25,787 bp of inverted repeat, including 130 genes (85 protein-coding genes, 8 rRNAs, and 37 tRNAs). Phylogenetic trees show that H. corniculata is a sister to S. bimaculata, nested within Swertia, consistent with previous phylogenetic studies.
Halenia Borkh. (Gentianaceae) consists of approximately 40 annual or perennial species distributed in eastern Europe, East Asia, and the Americas (von Hagen and Kadereit Citation2003). Halenia is the only member in Gentianaceae that has spurred corollas, and it is shown as monophyletic within the family (von Hagen and Kadereit Citation2002). Halenia species have been used as herbal medicine sources to treat hepatic diseases, such as hepatitis and jaundice, in East Asia (Yang et al. Citation1991). Halenia corniculata (L.) Cornaz, distributed in northeastern China, Mongolia, Korea, northern Japan, and the Russian Far East, is one of two Asian species and is distinguished from the Himalayan H. elliptica D. Don by having yellow corollas and linear-lanceolate calyxes (Toyokuni and Yamazaki Citation1993; Ho and Pringle Citation1995; Park Citation2007). H. corniculata is listed as an endangered species in Korea by the Ministry of the Environment of Korea (http://species.nibr.go.kr). Only a few populations with less than 100 individuals have been reported in Korea. These populations, which can be found in open meadow-like habitats at high elevations on mountains, are threatened by habitat destruction and over-collection.
To understand its genetic background, we determined its complete chloroplast genome. The total DNA of H. corniculata isolated from Mt. Hwaaksan, Korea (Voucher in the Herbarium of the National Institute of Biological Resources, Korea (KB); [Oh 7377]) was extracted from fresh leaves using a DNeasy Plant Mini Kit (QIAGEN, Hilden, Germany). The genome was sequenced using HiSeq4000 of Macrogen Inc., Korea, and de novo assembly and confirmation were performed with Velvet 1.2.10 (Zerbino and Birney Citation2008), SOAP GapCloser 1.12 (Zhao et al. Citation2011), BWA 0.7.17 (Li Citation2013), and SAMtools 1.9 (Li et al. Citation2009). Geneious R11 11.0.5 (Biomatters Ltd., Auckland, New Zealand) was used for annotation based on the Swertia mussotii chloroplast genome (NC031155; Xiang et al. Citation2016).
The chloroplast genome of H. corniculata (GenBank accession is MK606372) is 153,198 bp (GC ratio is 38.2%) and has four subregions: 83,252 bp of large-single-copy (LSC: 36.4%) and 18,372 bp of small-single-copy (SSC: 32.2%) regions are separated by 25,787 bp of an inverted repeat (IR: 43.4%). It contains 130 genes (85 protein-coding genes, 8 rRNAs, and 37 tRNAs); 19 genes (8 protein-coding gene, 4 rRNAs, and 7 tRNAs) are duplicated in the IR regions.
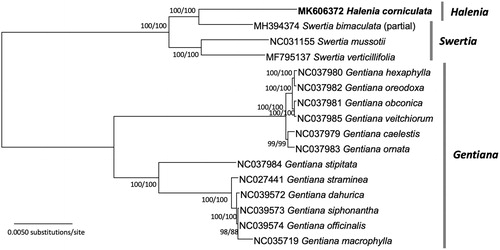
Complete or partial chloroplast genomes from H. corniculata, 3 Swertia chloroplast genomes, and 12 Gentiana genomes as outgroups were aligned using MAFFT 7.388 (Katoh and Standley Citation2013). Phylogenetic trees using maximum likelihood and neighbor-joining methods were drawn by MEGA X (Kumar et al. Citation2018). The phylogenetic trees show that H. corniculata is a sister to S. bimaculata, nested within Swertia (), in good agreement with a previous phylogenetic analysis of Swertiinae (von Hagen and Kadereit Citation2002). In addition, two major clades in Gentiana correspond to the findings of a previous phylogeny study (Mishiba et al. Citation2009). The sample of H. corniculata in this study, which represents the southernmost population of the species, will contribute to a better understanding of its haplotype diversity and conservation biology.
Figure 1. A Neighbor-joining tree (with 10,000 bootstrap replicates) of Halenia, Swertia, and Gentiana chloroplast genomes from Gentianaceae: H. corniculata (MK606372 in this study), S. bimaculata (MH394374), S. mussotii (NC031155), S. verticillifolia (MF795137), G. macrophylla (NC035719), G. straminea (NC027441), G. obconica (NC037981), G. oreodoxa (NC037982), G. veitchiorum (NC037985), G. ornata (NC037983), G. caelestis (NC037979), G. hexaphylla (NC037980), G. stipitata (NC037894), G. officinalis (NC039574), G. siphonantha (NC039573), and G. dahurica (NC039572). Phylogenetic tree based on maximum likelihood (with 1000 replicates) produced a tree nearly identical to the NJ tree. The numbers below the branches indicate the corresponding bootstrap support values from the neighbor joining and maximum likelihood methods.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Ho T, Pringle J. 1995. Gentianaceae. Flora of China, Vol. 16. Beijing and St. Louis: Science Press and Missouri Botanical Garden.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol Biol Evol. 35:1547–1549.
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The sequence alignment/map format and SAMtools. Bioinformatics. 25:2078–2079.
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv Preprint arXiv. 13033997.
- Mishiba K-i, Yamane K, Nakatsuka T, Nakano Y, Yamamura S, Abe J, Kawamura H, Takahata Y, Nishihara M. 2009. Genetic relationships in the genus Gentiana based on chloroplast DNA sequence data and nuclear DNA content. Breeding Science. 59:119–127.
- Park C-W. 2007. Gentianaceae. The genera of vascular plants of Korea. Seoul: Academy Publ.
- Toyokuni H, Yamazaki T. 1993. Gentianaceae. Flora of Japan, Vol. 3a. Tokyo: Kodansha.
- von Hagen KB, Kadereit JW. 2002. Phylogeny and flower evolution of the Swertiinae (Gentianaceae-Gentianeae): Homoplasy and the principle of variable proportions. Syst Bot. 27:548–573.
- von Hagen KB, Kadereit JW. 2003. The diversification of Halenia (Gentianaceae): ecological opportunity versus key innovation. Evolution. 57:2507–2518.
- Xiang B, Li X, Qian J, Wang L, Ma L, Tian X, Wang Y. 2016. The complete chloroplast genome sequence of the medicinal plant Swertia mussotii using the PacBio RS II platform. Molecules. 21:1029.
- Yang Y, Ho T, Lu S, Huang R, Wang Z. 1991. Tibetan medicines. Xining: Qinghai People’s Press.
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18:821–829.
- Zhao Q-Y, Wang Y, Kong Y-M, Luo D, Li X, Hao P. 2011. Optimizing de novo transcriptome assembly from short-read RNA-Seq data: a comparative study. BMC Bioinformatics. 12:S2.
