Abstract
Bolivia is believed to be the origin of the Capsicum eximium, a wild chilli pepper. In this study, we sequenced the complete chloroplast (cp) genome sequence of C. eximium to investigate its phylogenetic relationship in the family Solanaceae. The complete cp genome sequence is 156,947 bp in length with 37.7% overall GC content and exhibits a typical quadripartite structure comprising one pair of inverted repeats (25,847 bp) separated by a small single-copy region (17,912 bp) and a large single-copy region (87,341 bp). The cp genome contains 113 unique genes, including 79 protein coding genes, 30 tRNA genes, and 4 rRNA genes. Of these, 21 genes are duplicated in the inverted repeat regions. The phylogenetic analysis indicated C. eximium is clustered in the Capsicum clade.
Introduction
Pepper (Capsicum spp.) is native to tropical America; however, due to dispersal through humans since prehistoric times the original geographic origin and distribution were difficult to determine. Of the 27 recognized Capsicum species, 17 species have been reported ranges with Bolivia; in which C. eximium is believed to be found only in Bolivia and northern Argentina (Eshbaugh Citation1975). Recent research on wild Capsicum species clearly noticed an increased interest in this genus, especially in Bolivia; though, detailed descriptions are still limited or totally absent (Avila et al. Citation2012). In order to conserve the genetic diversity, a large number of wild species have been collected and established with genebanks worldwide, which is an essential prerequisite for developing new cultivars with desirable agronomic traits.
Understanding the germplasm diversity is vital for efficient transfer of valuable traits from wild species to cultivar for developing ecologically and economically sustainable cultivar varieties. Nowadays, the whole genome sequencing approaches are considered as cost-effective and used to understand the genetic diversity of wild species (Liu et al. Citation2016; Park Citation2016, Citation2017; Tsuruta et al. Citation2017). Chloroplast genome sequence of various Capsicum species was reported previously (Jo et al. Citation2011; Kim et al. Citation2016; Park et al. Citation2016; Shim et al. Citation2016). Here, we report the chloroplast genome sequence of Bolivian wild chilli pepper, C. eximium to find its internal relationships within the family Solanaceae.
Genomic DNA sample of Bolivian wild chili pepper, C. eximium (Accession No. K153259) was obtained from the National Agrobiodiversity Center, Republic of Korea. Total genomic DNA of the 40-day-old plant was used to decode the genome by Illumina MiSeq sequencing platform (http://www.Lab.genomics.com/kor/) with pair-end (2 × 300 bp) library. A total of 5,670,734 clean reads were obtained from 9,145,294 raw reads and mapped with the reference cp genome, C. annuum (JX270811), which contains 108,396 aligned reads with about an average 144× coverage. The cpDNA was annotated by the Dual Organellar Genome Annotator (DOGMA; http://dogma.ccbb.utexas.edu/) and tRNAscan-SE (Lowe and Chan Citation2016).
The cp genome sequence with complete annotation information was deposited to NCBI GenBank under the accession number KX913220. The total length of the chloroplast genome is 156,947 bp, with 37.7% overall GC content. With typical angiosperm plastome structure, a pair of IRs (inverted repeats) of 25,847 bp was separated by a small single copy (SSC) region of 17,912 bp and a large single copy (LSC) region of 87,341 bp. The cp genome contains 113 unique genes, including 79 protein coding genes, 30 tRNA genes, and 4 rRNA genes. Of these, 21 genes are duplicated in the inverted repeat regions, 15 genes, and 6 tRNA genes contain one intron, while two genes (ycf3 and rps12) have two introns.
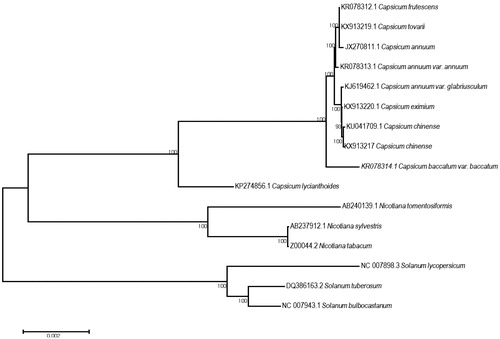
To further evaluate the phylogenetic position, the cp genome sequences of Solanaceae species were downloaded from the NCBI database (). Here, we aligned all the 16 cp genome sequences by MAFFT v7.304 (Katoh and Standley Citation2016) and constructed a maximum likelihood (ML) tree with 1000 bootstrap replicates using MEGA6 (Tamura et al. Citation2013) software. Our results showed that C. eximium was clustered in the Capsicum clade. The phylogenomic analyses of Solanaceae cp genomes were consistent with a previous study (D'Agostino et al. Citation2018).
Disclosure statement
The authors are highly grateful to the published genome data in the public database. The authors declare no conflicts of interest and are responsible for the content.
Additional information
Funding
References
- Avila T, Atahuachi M, Reyes X, Claure T, Zonneveld MV. 2012. Collection, taxonomic identification and cropping methodologies development for some species of Capsicum in Bolivia. Paper presented at The 21st International Pepper Conference; November 4–6; Florida, USA.
- D'Agostino N, Tamburino R, Cantarella C, De Carluccio V, Sannino L, Cozzolino S, Cardi T, Scotti N. 2018. the complete plastome sequences of eleven capsicum genotypes: insights into DNA variation and molecular evolution. Genes. 9:503.
- Eshbaugh WH. 1975. Genetic and biochemical systematic studies of chili peppers (Capsicum- Solanaceae). Bull Torrey Botanic Club. 102:396–403.
- Jo YD, Park J, Kim J, Song W, Hur CG, Lee YH, Kang BC. 2011. Complete sequencing and comparative analyses of the pepper (Capsicum annuum L.) plastome revealed high frequency of tandem repeats and large insertion/deletions on pepper plastome. Plant Cell Rep. 30:217–229.
- Katoh K, Standley DM. 2016. A simple method to control over-alignment in the MAFFT multiple sequence alignment program. Bioinformatics (Oxford, England). 32:1933–1942.
- Kim T-S, Lee J-R, Raveendar S, Lee G-A, Jeon Y-A, Lee H-S, Ma K-H, Lee S-Y, Chung J-W. 2016. Complete chloroplast genome sequence of Capsicum baccatum var. baccatum. Mol Breed. 36:1–5.
- Liu F, Tembrock LR, Sun C, Han G, Guo C, Wu Z. 2016. The complete plastid genome of the wild rice species Oryza brachyantha (Poaceae). Mitochondrial DNA Part B. 1:218–219.
- Lowe TM, Chan PP. 2016. tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44:W54–W57.
- Park T-H. 2016. The complete chloroplast genome sequence of potato wild relative species, Solanum nigrum. Mitochondrial DNA Part B. 1:858–859.
- Park T-H. 2017. The complete chloroplast genome of Solanum berthaultii, one of the potato wild relative species. Mitochondrial DNA Part B. 2:88–89.
- Park H-S, Lee J, Lee S-C, Yang T-J, Yoon JB. 2016. The complete chloroplast genome sequence of Capsicum chinense Jacq. (Solanaceae). Mitochondrial DNA Part B. 1:164–165.
- Shim D, Raveendar S, Lee JR, Lee GA, Ro NY, Jeon YA, Cho GT, Lee HS, Ma KH, Chung JW. 2016. The complete chloroplast genome of Capsicum frutescens (Solanaceae). Appl Plant Sci. 4.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Bio Evol. 30:2725–2729.
- Tsuruta S-i, Ebina M, Kobayashi M, Takahashi W. 2017. Complete chloroplast genomes of Erianthus arundinaceus and Miscanthus sinensis: Comparative genomics and evolution of the Saccharum complex. PLoS One. 12:e0169992.