Abstract
In this study, the complete mitochondrial genome of the Neverita didyma (Mollusca: Gastropoda) was sequenced. The mitochondrial genome is 15,252 bp in length, containing 13 protein-coding genes, 2 ribosomal RNA genes, 22 transfer RNA genes. The overall nucleotides base composition of the heavy strand is 30.47% for A, 14.09%% for C, 15.89%for G, and 39.56% for T with an AT-rich feature (70.03%). All the protein-coding genes begin with an ATG codon, the common termination codon is TAA, except for nd2 and nd4l terminated with TAG and nd5 terminated by incomplete codon TA. Seen from the phylogenetic tree, Naticarius hebraeus, and N. didyma from the same Superfamily (Naticoidea) clustered into one branch. The data would provide useful data for further studies on population genetics and molecular systematics.
Neverita didyma is common in the Yellow Sea Ecoregion. In recent years, the wild stocks of N. didyma have experienced dramatic population declines due to over-exploitation and the deterioration of the coastal environment (Li et al. Citation2012; Liu et al. Citation2013). To prevent the over-exploitation of this species, genetic information is needed and helpful for rational utilization of N. didyma resources. In this paper, we present the complete mitochondrial genome of N. didyma by using the next-generation sequencing (NGS) techniques strategy (Wang et al. Citation2015), which can help to better understand the phylogenetic status of this species in Gastropoda. The N. didyma specimen used in this study was collected from Jiaozhou Bay (36.11°N, 120.19°E) during October 2018. All examined specimens were preserved at the Marine Ecology Research Center of the First Institute Oceanography, MNR in Qingdao. The complete mitochondrial genome of N. didyma (Genbank accession no MK548644) is 15,252 bp in length, containing 13 protein-coding genes, 2 ribosomal RNA genes (12S rRNA and 16S rRNA), 22 transfer RNA genes (tRNA), which is the same as the other Naticoidea species, Naticarius hebraeus (David et al. Citation2015). The CDS sequences correspond to the genes cox1, cox2, cox3, cytb, atp6, atp8, nd1, nd2, nd3, nd4, nd4l, nd5, nd6. All these genes are in the heavy strand. The rRNA genes, 12S rRNA, and 16S rRNA are also present in the heavy strand. The 22 tRNA genes are distributed in both the strands. The overall nucleotides base composition of the heavy strand is 30.47% for A, 14.09% for C, 15.89% for G, and 39.56% for T with an AT-rich feature (70.03%). All the protein-coding genes begin with an ATG codon, the common termination codon is TAA, except for nd2 and nd4l terminated with TAG, and nd5 terminated by incomplete codon TA. The longest gene is nd5 (1718 bp) among the protein-coding genes, whereas the shortest is the atp8 gene (159 bp). The two ribosomal RNA genes, 12S rRNA (881 bp), and 16S rRNA (1301 bp) are located between the tRNA-Glu(TTG) and tRNA-Leu(TAA) genes and are further separated by the tRNA-Val(TAC) gene.
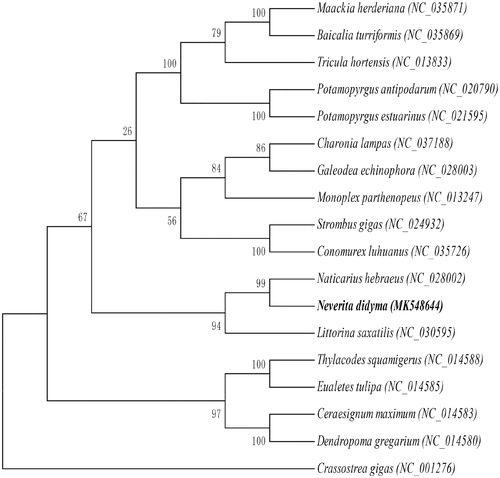
The maximum likelihood tree (by RAxML 8.1.5, Heidelberg, Germany) (Alexandros Citation2006). based on mitochondrial genome nucleotide sequences of the N. didyma and the other 17 species of Mollusca were constructed under the mutation model of GTR + I+G (by jModelTest 2.1.7, Vigo, Spain) (David Citation2008). Seen from the phylogenetic tree, Naticarius hebraeu and N. didyma from the same Superfamily (Naticoidea) clustered into one branch, and N. didyma has a more close relationship with N. hebraeus than other species .
Figure 1. Maximum likelihood tree based on mitochondrial genome nucleotide sequences of the Neverita didyma (MK548644) and the other 17 species of Mollusca under the mutation model of GTR + I+G. Genbank accession numbers of the sequences were used for the tree as follows: Naticarius hebraeus (NC_028002.1), Charonia lampas (NC_037188.1), Baicalia turriformis (NC_035869.1), Maackia herderiana (NC_035871.1), Conomurex luhuanus (NC_035726.1), Galeodea echinophora (NC_028003.1), Ceraesignum maximum (NC_014583.1), Potamopyrgus estuarinus (NC_021595.1), Potamopyrgus antipodarum (NC_020790.1), Littorina saxatilis (NC_030595.1), Strombus gigas (NC_024932.1), Thylacodes squamigerus (NC_014588.1), Eualetes tulipa (NC_014585.1), Dendropoma gregarium (NC_014580.1), Tricula hortensis (NC_013833.1), Monoplex parthenopeus (NC_013247.1), Crassostrea gigas (NC_001276.1).
Disclosure statement
The authors report that they have no conflicts of interest. The authors alone are responsible for the content and writing of this article.
Additional information
Funding
References
- Alexandros S. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 22:2688–2690.
- David O, José T, Rafael Z. 2015. Caenogastropod mitogenomics. Mol Phylogenet Evol. 93:118–128.
- David P. 2008. jModelTest: phylogenetic model averaging. Mol Biol Evol. 25:1253–1256.
- Li N, Li Q, Jin MY, Kong LF, Yu H. 2012. Isolation and characterization of 20 microsatellite loci in Neverita didyma (Roding 1798). Conservation Genet Resour. 4:479–481.
- Liu H, Xu M, Wu C. 2013. Evaluation of nutritional composition in Nevertia didyma and Natica vitellus from Zhoushan Sea area. Food Sci. 34:228–231.
- Wang Q, Wang J, Luo J, Chen GH. 2015. The complete mitochondrial genome of the Synanceia verrucosa (Scorpaeniformes: Synanceiidae). Mitochondrial DNA. 26:1–2.
