Abstract
Acerina cernua is a specific percid species that naturally occurs in Xinjiang Irtysh River basin, China. The complete mitochondrial genome of the ruffe A. cernua was first determined (Accession number MK509809), with a total length of 16,677 nucleotides, of which 15,579 nucleotides are coding DNA, and 1098 nucleotides are non-coding DNA. It has the common feature with those of other fishes with respect to genome structure and gene arrangement. It contained 13 protein-coding genes, two ribosomal RNA genes, 22 transfer RNA genes, and one control region (D-loop region). The complete mitochondrial genome of ruffe A. cernua provides an important data set for further study on its classification.
Keywords:
Acerina cernua, which belongs to Acerina Cuvier, Percidae, Perciformes, Actinopterygii, Osteichthyes, is a specific percid species that naturally occurs in Xinjiang Irtysh River basin, China (Zhu et al. Citation2014). However, there is still no study on the complete mtDNA sequences for A. cernua has been reported. So, it is important to obtain the whole mitogenome of A. cernua for the further study. The specimen of A. cernua, named as Acernua-01 based on the phylogenetic analysis, was collected from Irtysh River, Altay Prefecture, China (47°83′N, 88°13′E) in 2014 and stored in College of Life Sciences, Linyi University, Linyi, China. Genomic DNA was extracted from A. cernua muscle according to Liu et al. (Citation2012, Citation2014). The complete mitochondrial genome was sequenced using a shotgun approach and assembly. Protein-coding genes were analyzed by ORF Finder (http://www.ncbi.nlm.nih.gov/gorf/gorf.html) using the invertebrate mitochondrial code. The tRNA genes were identified using ARWEN (Laslett and Canback Citation2008) and tRNA-scan SE (Lowe and Eddy Citation1997).
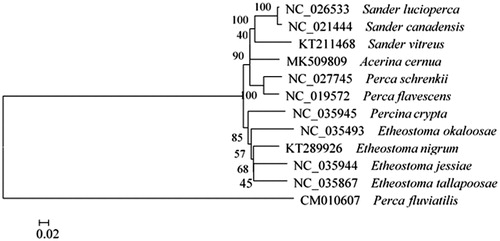
The complete A. cernua mitochondrial genome (Accession number MK509809) was 16,677 nucleotides long, of which 15,579 nucleotides are coding DNA, and 1098 nucleotides are non-coding DNA. The mitochondrial genome of A. cernua was found to contain 13 protein-coding genes, two ribosomal RNA genes, 22 transfer RNA genes, and one control region. Its arrangement and composition is also comparable to other vertebrates (Liang et al. Citation2012; Liu et al. Citation2013; Xu et al. Citation2015; Balakirev et al. Citation2016; Shan and Liu Citation2016). A phylogenetic tree was constructed based on the comparison of the complete mitochondrial genome sequences with other Percidae species using Neighbour-Joining method (). There were 13 intergenic spacers (total 84 bp) and 11 overlapping regions (total 34 bp) among the genes. The overall base composition was 27.85% A, 27.58% T, 27.90% C, and 16.67% G, with an AT content of 55.43%. To investigate the nucleotide bias, skew for a given strand was calculated as (A–T)/(A + T) or (G–C)/(G + C) (Perna and Kocher Citation1995). The AT and GC skews for the A. cernua mitochondrial genome were 0.005 and −0.252, respectively; this finding indicated that the strand that encoded genes contained more A and C than T and G, and this skew was evidence of codon usage bias. All 13 protein-coding genes identified in the A. cernua mitochondrial genome, started with the orthodox initiation codon ATG, except for COX1, which began with GTG. The ORFs of six protein-coding genes, ND1, ND2, COX1, ATP8, ATP6, COX3, ND4L, and ND5, ended with a TAA codon, and the remaining genes ended with incomplete stop codon T or AT. The length of 12S rRNA and 16S rRNA was 949 bp and 1691 bp, respectively. The intergenic region between 12S rRNA and 16S rRNA was found to be 72 bp and to contain only one tRNA (tRNA-val). The lengths of 22 tRNA coding genes range in size from 66 to 74 nucleotides. The control region was located between tRNA-pro and tRNA-phe with a length of 1014 bp. The data would facilitate further investigations of phylogenetic relationships within Percidae.
Figure 1. A phylogenetic tree constructed based on the comparison of complete mitochondrial genome sequences of the Percidae, A. cernua and other twelve species of Percidae family. They are Perca fluviatilis (European perch), P. schrenkii (balkhash perch), P. flavescens (yellow perch), Etheostoma okaloosae (okaloosa darter), E. nigrum (johnny darter), E. jessiae (blueside darter), E. tallapoosae (tallapoosa darter), Percina crypta (halloween darter), Sander vitreu (walleye), S. lucioperca (pike-perch), and S. canadensis (sauger). Genbank accession numbers for all sequences are listed in the figure. The numbers at the nodes are bootstrap percent probability values based on 1000 replications.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the article.
Additional information
Funding
References
- Balakirev ES, Romanov NS, Ayala FJ. 2016. Complete mitochondrial genome of the yellow-spotted grayling Thymallus flavomaculatus (Salmoniformes, Salmonidae). Mitochondrial DNA Part B. 1:289–290.
- Laslett D, Canback B. 2008. ARWEN: a program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics. 24:172–175.
- Liang HW, Hu GF, Li Z, Zou GW, Liu XL. 2012. Mitochondrial DNA sequence of yellow catfish (Pelteobagrus fulvidraco). Mitochondrial DNA. 23:170–172.
- Liu YG, Guo YG, Hao J, Liu LX. 2012. Genetic diversity of swimming crab (Portunus trituberculatus) populations from Shandong peninsula as assessed by microsatellite markers. Biochem Syst Ecol. 41:91–97.
- Liu YG, Kurokawa T, Sekino M, Tanabe T, Watanabe K. 2013. Complete mitochondrial DNA sequence of the ark shell Scapharca broughtonii: an ultra-large metazoan mitochondrial genome. Comp Biochem Phys D. 8:72–81.
- Liu YG, Liu LX, Xing SC. 2014. Development and characterization of thirteen polymorphic microsatellite loci in the Cynoglossus semilaevis. Conserv Genet Resour. 6:683–684.
- Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955–964.
- Perna NT, Kocher TD. 1995. Patterns of nucleotide composition at fourfold degenerate sites of animal mitochondrial genomes. J Mol Evol. 41:353–358.
- Shan WJ, Liu YG. 2016. The complete mitochondrial DNA sequence of the cape hare Lepus capensis pamirensis. Mitochondrial DNA A DNA Mapp Seq Anal. 27:4572–4573.
- Xu D, He CQ, Li QH, He J, Ma HM. 2015. The complete mitochondrial genome of the Daweizi pig. Mitochondrial DNA. 26:640–641.
- Zhu MY, Wang X, Zhou Y, Song Y, Qin L, Hao CL, Zhang F, Yue C. 2014. Population dynamics of diplostomum infectionin Acerina cernua in Ergis River. J Hydroecol. 35:78–81.
