Abstract
In this study, the complete mitochondrial genome (mitogenome) sequence of Platycephalus sp.1 was sequenced by long polymerase chain (PCR) reaction and primer walking method. The mitogenome was total 16552 base pairs (bp) in length and contained 2 ribosomal RNA (rRNA), 13 protein-coding genes (PCGs), 22 transfer RNA (tRNA), and 1 non-coding region (control region) as those found in other vertebrates. The overall base composition was displayed as 25.8% T, 30.0% C, 27.0% A, and 17.2% G. The neighbour-joining tree was reconstructed based on the 13 PCGs sequences of Platycephalus sp.1 and other six species. The mitogenome data would provide useful information for further studies on the taxonomy and phylogenetic analysis of Platycephalidae.
Platycephalus sp.1 (Nakabo Citation2002) has been long recognized in the waters around Japan (Kamei and Ishiyama Citation1969), which differs morphologically from other congeners of genus Platycephalus (Masuda et al. Citation1997; Yamada et al. Citation2007). Subsequently, a new record of Platycephalus sp.1 from China was documented based on morphological characters and DNA barcoding by Qin et al. (Citation2013). This study expanded the distribution range of Platycephalus sp.1 and suggested that it was widely distributed in coastal waters of China. There was no specific scientific name of Platycephalus sp.1. Therefore, it was necessary for further studies on Platycephalus sp.1. The complete mitogenome of Platycephalus sp.1 in this study would facilitate further taxonomy and phylogenetic analysis.
The specimen of Platycephalus sp.1 was collected from the coast of Qingdao, China (120.4°E, 35.9°N) in 2011 and preserved in absolute ethanol at the Fishery Ecology & Marine Biodiversity Laboratory, Fishery College, Zhejiang Ocean University (Zhoushan). Morphological identification was conducted according to Masuda et al. (Citation1997) and Yamada et al. (Citation2007). Total genomic DNA was extracted by proteinase K digestion followed by a standard phenol-chloroform extraction method. The complete mitogenome sequence was determined by long polymerase chain reaction (PCR) and primer walking method (Cheng et al. Citation1994). The assembled mitogenome was manually checked and deposited in GenBank with accession number MK344191.
The complete mitogenome of Platycephalus sp.1 was 16552 bp in length. It consisted of 2 rRNA genes, 13 protein-coding genes (PCGs), 22 transfer RNA (tRNA) genes, and the control region. Gene arrangement was in accordance with the canonical structure of vertebrates’ mitogenome. Thirteen coding genes of mitogenome were located on H-strand with the exception of ND6, and eight tRNA genes were transcribed from L-strand. The overall base composition was displayed as 25.8% T, 30.0% C, 27.0% A, and 17.2% G, with a slight excess of AT and anti-G bias as observed in most fishes (Miya et al. Citation2003). The 13 protein-coding genes were totally 11400 bp in size, accounting for 68.9% of the whole mitogenome. Twenty-two tRNA genes ranged in size from 68 to 74 bp. The mitogenome of Platycephalus sp.1 also contained a small subunit rRNA (12S rRNA) and a large subunit rRNA (16S rRNA), which were 951 and 1695 bp in length, respectively. The termination-associated motif sequence (TAS), central conserved domain (CSB-F, CSB-E, and CSB-D), and conserved sequence domain (CSB-1, CSB-2, and CSB-3) were recognized in control region with a total length of 1224 bp.
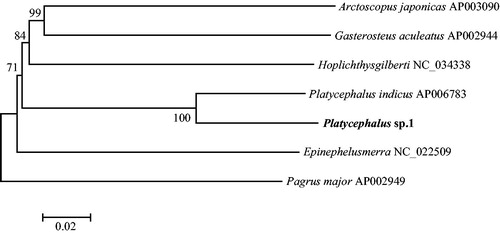
On account of only one available complete mitogenome of family Platycephalidae in the GenBank database, other six applicable complete mitogenomes according to Miya et al. (Citation2013) were retrieved to estimate the phylogenetic position of Platycephalus sp.1. The neighbour-joining (NJ) phylogenetic tree was reconstructed by MEGA version 6.06 (Tamura et al. Citation2013) based on the concatenated nucleotide sequences of 13 PCGs. It could be easily distinguished that Platycephalus sp.1 clustered with Platycephalus indicus with the high support values ().
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Cheng S, Chang SY, Gravitt P, Respess R. 1994. Long PCR. Nature. 369:684–685.
- Kamei M, Ishiyama R. 1969. Morphology and ecology of two types of flathead fishes belonging to the genus Platycephalus. Proc MeetJapanSocSciFish. Tokyo, Japan.
- Masuda Y, Haraguchi M, Ozawa T, Matsui S, Hayashi S. 1997. Morphological and biochemical comparisons of two flathead species of the genus Platycephalus collected from the Western Suonada and Southern Yatsushiro Seas, Japan. Nippon Suisan Gakk. 3:345–352.
- Miya M, Friedman M, Satoh TP, Takeshima H, Sado T, Iwasaki W, Yamanoue Y, Nakatani M, Mabuchi K, Inoue JG, et al. 2013. Evolutionary origin of the scombridae (tunas and mackerels): members of a paleogene adaptive radiation with 14 other pelagic fish families. PLoS One. 8:e73535.
- Miya M, Takeshima H, Endo H, Ishiguro NB, Inoue JG, Mukai T, Satoh TP, Yamaguchi M, Kawaguchi A, Mabuchi K, et al. 2003. Major patterns of higher teleostean phylogenies: a new perspective based on 100 complete mitochondrial DNA sequences. Mol Phylogenet Evol. 26:121–138.
- Nakabo T. 2002. Fishes of Japan with pictorial keys to the species. Kanagawa, Japan: Tokai University Press.
- Qin Y, Song N, Zou JW, Zhang ZH, Cheng GP, Gao TX, Zhang XM. 2013. A new record of a flathead fish (Teleostei: Platycephalidae) from China based on morphological characters and DNA barcoding. Chin J Ocean Limnol. 31:617–624.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- Yamada U, Tokimura M, Horikawa H, Nakabo T. 2007. Fishes and fisheries of the East China and yellow seas. Kanagawa, Japan: Tokai University Press.