Abstract
We report the complete mitogenome of a rough-toothed dolphin, Steno bredanensis CRI008296, stranded in Korea. Overall, structure and taxonomical position of the mitogenome were analysed, and its determined haplotype indicated that the same subpopulation is distributed between Korea and Japan.
The rough-toothed dolphin, Steno bredanensis (Lesson, 1828), is the sole species in the genus Steno widely distributed in tropical to subtropical regions of all three major oceans (Miyazaki and Perrin Citation1994; West et al. Citation2011). Currently, this species is listed in Appendix II of the Convention on International Trade in Endangered Species (CITES Citation2017) and categorized as ‘Least Concern’ species by the International Union for Conservation of Nature and Natural Resources Red List of Threatened Species (Hammond et al. Citation2012). Although several subpopulations of this species were genetically differentiated (Oremus et al. Citation2012; da Silva et al. Citation2015; Kerem et al. Citation2016; Albertson et al. Citation2017), the molecular diversity of the West Pacific subpopulation is not well understood and its presence in South Korea remains uncertain (Hammond et al. Citation2012). Since 2016, we have investigated the biological diversity of endangered marine animals present in Korean coastal waters for their conservation (Lee et al. Citation2018). As part of that project, we determined the haplotype and complete mitogenome of a S. bredanensis individual stranded in Korea.
Muscle tissue was collected from the carcass of an adult male S. bredanensis (voucher no. CRI008296 deposited in the Cetacean Research Institute, National Institute of Fisheries Science) that was stranded at the seashore (34°43′36′′N 127°45′53″E) in Yeosu (South Korea) in April 2018. The S. bredanensis mitogenome was obtained using PCR amplifications and assembled, and annotated as previously described (Kim et al. Citation2017; Lee et al. Citation2018).
The S. bredanensis mitogenome (MH910343) was 16,386 bp long (61.2% A + T content) and included a typical set of 37 genes (two rRNAs, 22 tRNAs, and 13 protein-coding genes) that were almost identical to those from other related Delphinidae species. The average mitogenomes nucleotide identity between S. bredanensis and other Delphinidae species was determined as previously described (Lee et al. Citation2018). A S. bredanensis individual from the Atlantic Ocean subpopulation (JF681038.1) was most similar to CRI008296 (99.1% similarity), the highest species-specific value was obtained for Stenella attenuata (NC_012051, 94.4%), and the lowest similarity was with Orcinus orca (NC_023889, 92.9%). To determine its haplotype, the D-loop sequence of CRI008296 was compared with those from other S. bredanensis individuals in the GenBank database. The results verified that the haplotype of CRI008296 was h7, which was collected from Japan (Oremus et al. Citation2012), suggesting that the same subpopulation is distributed between Korea and Japan.
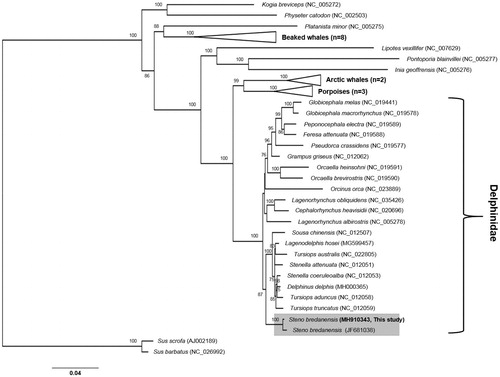
The taxonomical position of CRI008296 was determined using multigene phylogenetic analysis (Kim et al. Citation2017). The resultant phylogeny clustered CRI008296 with other S. bredanensis (JF681038.1) but separate from the other genus within Delphinidae, as previously reported (Cunha et al. Citation2011) (). The mitogenomic information of S. bredanensis stranded in Korea might provide important insights into the genetic diversity of its population structure and conservation.
Figure 1. Maximum-likelihood phylogeny based on the 13 concatenated protein-coding genes from the available Odontoceti mitogenomes. Numbers at the branches indicate bootstrapping values obtained with 1000 replicates, and only bootstrap values >70% are indicated. The scale bar represents 0.04 nucleotide substitutions per site.
Disclosure statement
The authors declare that they have no conflict of interest.
Additional information
Funding
References
- Albertson GR, Baird RW, Oremus M, Poole MM, Martien KK, Baker CS. 2017. Staying close to home? Genetic differentiation of rough-toothed dolphins near oceanic islands in the central Pacific Ocean. Conserv Genet. 18:33–51.
- [CITES] Convention on International Trade in Endangered Species of Wild Fauna and Flora. 2017. Appendices I, II, and III. [accessed 2019 Feb 14]. https://www.cites.org/eng/app/appendices.php.
- Cunha HA, Moraes LC, Medeiros BV, Lailson-Brito J, Jr, da Silva VMF, Solé-Cava AM, Schrago CG. 2011. Phylogenetic status and timescale for the diversification of Steno and Sotalia dolphins. PLoS One. 6:e28297.
- da Silva DMP, Azevedo AF, Secchi ER, Barbosa LA, Flores PAC, Carvalho RR, Bisi TL, Lailson-Brito J, Cunha HA. 2015. Molecular taxonomy and population structure of the rough-toothed dolphin Steno bredanensis (Cetartiodactyla: Delphinidae). Zool J Linnean Soc. 175:949–962.
- Hammond PS, Bearzi G, Bjørge A, Forney KA, Karkzmarski L, Kasuya T, Perrin WF, Scott MD, Wang JY, Wells RS, et al. 2012. Steno bredanensis. The IUCN red list of threatened species 2012. e.T20738A17845477. [accessed 2019 Feb 14]. http://dx.doi.org/10.2305/IUCN.UK.2012.RLTS.T20738A17845477.en.
- Kerem D, Goffman O, Elasar M, Hadar N, Scheinin A, Lewis T. 2016. The rough-toothed dolphin, Steno bredanensis, in the eastern Mediterranean sea: a relict population? Adv Mar Biol. 75:233–258.
- Kim JH, Lee YR, Koh JR, Jun JW, Giri SS, Kim HJ, Chi C, Yun S, Kim SG, Kim SW, et al. 2017. Complete mitochondrial genome of the beluga whale Delphinapterus leucas (Cetacea: Monodontidae). Conserv Genet Resour. 9:435–438.
- Lee K, Lee JMO, Sohn H, Cho Y, Choi YM, Kim HK, Kim JH, Jeong DG. 2018. Complete mitochondrial genome of the Pacific white-sided dolphin Lagenorhynchus obliquidens (Cetacea: Delphinidae). Conserv Genet Resour. 10:201–204.
- Miyazaki N, Perrin WF. 1994. Rough-toothed dolphin Steno bredanensis (Lesson, 1828). In: Ridgway SH, Harrison R, editors. Handbook of marine mammals. Vol. 5. San Jose (CA): Academic Press; p. 1–21.
- Oremus M, Poole MM, Albertson GR, Baker CS. 2012. Pelagic or insular? Genetic differentiation of rough-toothed dolphins in the Society Islands, French Polynesia. J Exp Mar Biol Ecol. 432:37–46.
- West KL, Mead JG, White W. 2011. Steno bredanensis (Cetacea: Delphinidade). Mamm Species. 43:177–189.
