Abstract
As a Korean endemic plant, Salix koriyanagi Kimura ex Goerz belongs to the genus Salix L. which is the largest genus of Salicaceae. In this study, we presented a complete chloroplast genome from male individual of S. koriyanagi which is 155,548 bp and has four subregions: 84,415 bp of large single copy (LSC) and 16,125 bp of small single copy (SSC) regions are separated by 27,459 bp of inverted repeat (IR) regions including 131 genes (85 protein-coding gene, 8 rRNAs, and 38 tRNAs). The overall GC content of the chloroplast genome is 36.7% and those in the LSC, SSC, and IR regions are 34.4%, 31.0%, and 41.9%, respectively. SSC is inverted in comparison to that of female individual of S. koriyanagi. Phylogenetic trees show phylogenetic relationship of Salix and Populus genera.
Salix koriyanagi Kimura ex Goerz, belonging to Salix L. genus, largest genus in Salicaceae (Argus Citation1997), is a Korean endemic species. It is usually found nearby streams (Lee Citation2003). S. koriyanagi is dioecious species, separating male and female individuals (Argus Citation1997). In previous study of chloroplast genome of S. koriyanagi, female individual was selected (Kim J et al. Citation2019) to decipher its whole genome, addressing the next question: what the differences between male and female individuals in the aspect of genome sequences are.
To answer the question, total DNA was extracted from fresh leaves of S. koriyanagi male individual isolated from Cheonggyecheon (stream) in Seoul, Republic of Korea (Voucher was deposited in InfoBoss Cyber Herbarium (IN): Y. Kim, IB-00599) by using a DNeasy Plant Mini Kit (QIAGEN, Hilden, Germany). Genome sequencing was performed using HiSeq4000 (Illumina, San Diego, CA) at Macrogen, Korea. After filtering using Trimmomatic 0.33 (Bolger et al. Citation2014) and de novo assembly and confirmation were done by Velvet 1.2.10 (Zerbino and Birney Citation2008), SOAPGapCloser 1.12 (Zhao et al. Citation2011), BWA 0.7.17 (Li Citation2013), and SAMtools 1.9 (Li et al. Citation2009). Geneious R11 version 11.0.5 (Biomatters Ltd., Auckland, New Zealand) was used for annotation based on S. koriyanagi chloroplast complete genome (MK120982; Kim J et al. Citation2019).
The chloroplast genome of S. koriyanagi male individual (Genbank accession is MK541017; GC ratio is 36.7%) is 155,548 bp and has four subregions: 84,415 bp of large single copy (LSC; 34.4%) and 16,125 bp of small single copy (SSC; 31.0%) regions are separated by 27,549 bp of inverted repeat (IR; 41.9%). It contains 131 genes (85 protein-coding genes, 8 rRNAs, and 38 tRNAs); 19 genes (eight protein-coding genes, four rRNAs, and seven tRNAs) are duplicated in IR regions.
By comparing with S. koriyanagi female chloroplast genome (Kim J et al. Citation2019), SSC region is reversed, which is like many chloroplast genomes including Olea europaea subsp. europaea (Xia et al. Citation2016; Chen et al. Citation2017; Shen et al. Citation2017; Kim J et al. Citation2019; Kim Y et al. Citation2019; Min et al. Citation2019) . In addition, there is no sequence variation between male and female individuals because both individuals might be originated from the same parents or parents containing same chloroplast haplotype. It is similar case to those of Marchantia polymorpha subsp. rudalis (Kwon et al. Citation2019), Pseudostellaria longipedicellata, and Populus alba × P. glandulosa (Park et al. accepted) .
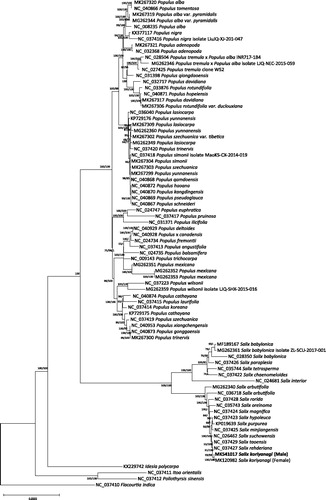
Sequence alignment of all available 80 complete chloroplast genomes from Salicaceae with correcting directions of SSC and LSC of six Salix genomes (Salix suchowensis, Salix purpurea, Salix arbutifolia, and Salix babylonica) was conducted by Multiple Alignment using Fast Fourier Transform (MAFFT 7.388) (Katoh and Standley Citation2013). Maximum likelihood (bootstrap repeat is 1,000) and neighbor joining (bootstrap repeat is 10,000) phylogenetic trees were constructed by IQ-TREE 1.6.6 (Nguyen et al. Citation2015) and MEGA X (Kumar et al. Citation2018), respectively. Phylogenetic trees present congruent with previous Salix (Lauron-Moreau et al. Citation2015; Wu Citation2015; Wu et al. Citation2015) as well as Populus phylogenetic analyses (Hamzeh and Dayanandan Citation2004; ), reflecting potential power of chloroplast genome for unrevealing high resolution phylogenetic relationships at genera level together with abundant chloroplast genomes of Salix genus.
Figure 1. Neighbor joining (bootstrap repeat is 10,000) and maximum likelihood (bootstrap repeat is 1,000) phylogenetic trees of 80 complete chloroplast genomes from Salicaceae: Salix koriyanagi (MK541017 in this study and MK120982), Salix suchowensis (NC_026462), Salix purpurea (KP019639), Salix rehderiana (NC_037427), Salix rorida (NC_037428), Salix taoensis (NC_037429), Salix tetrasperma (NC_035744), Salix paraplesia (NC_037426), Salix oreinoma (NC_035743), Salix minjiangensis (NC_037425), Salix magnifica (NC_037424), Salix interior (NC_024681), Salix hypoleuca (NC_037423), Salix chaenomeloides (NC_037422), Salix babylonica (NC_028350, MG262361, and MF189167), Salix arbutifolia (NC 036718 and MG262340), Populus yunnanensis (MG262360 and MK267299), Populus xiangchengensis (NC_040953), Populus × canadensis (NC_040928), Populus wilsonii (MG262359 and NC_037223), Populus trinervis (NC_037420 and MK267300), Populus trichocarpa (NC 009143), Populus tremula × Populus alba (MG262346 and NC_028504), Populus tremula (NC_027425), Populus tomentosa (NC_040866), Populus szechuanica var. tibetica (MK267302), Populus szechuanica (NC_037419 and MK267303), Populus simonii (NC_037418 and MK267304), Populus schneideri (NC_040867), Populus rotundifolia var. duclouxiana (MK267306), Populus rotundifolia (NC_033876), Populus qiongdaoensis (NC_031398), Populus qamdoensis (NC_040868), Populus pseudoglauca (NC_040869), Populus pruinosa (NC_037417), Populus nigra (NC_037416 and KX377117), Populus mexicana (MG262351, MG262352, and MG262353), Populus laurifolia (NC_037415), Populus lasiocarpa (NC_036040, MG262349, and MK267309), Populus koreana (NC_037414), Populus kangdingensis (NC_030870), Populus ilicifolia (NC_031371), Populus hopeiensis (NC_040871), Populus haoana (NC_040872), Populus gonggaensis (NC_040873), Populus fremontii (NC_024734), Populus euphratica (NC_024747), Populus deltoides (NC_040929), Populus davidiana (NC_032717 and MK267317), Populus cathayana (NC_040874), Populus balsamifera (NC_024735), Populus angustifolia (NC_037413), Populus alba var. pyramidalis (MG262344 and MK267319), Populus alba (NC_008235 and MK267320), Populus adenopoda (NC_032368 and MK267321), Populus cathayana (KP729175), Populus yunnanensis (KP729176), Poliothyrsis sinensis (NC_037412), Itoa orientalis (NC_037411), Idesia polycarpa (KX229742), and Flacourtia indica (NC_037410). Neighbor joining tree was used for displaying phylogenetic tree. The numbers above branches indicate bootstrap support values of neighbor joining maximum likelihood trees, respectively.
Disclosure statement
The authors declare that they have no competing interests.
Additional information
Funding
References
- Argus GW. 1997. Infrageneric classification of Salix (Salicaceae) in the new world. Syst Bot Monogr. 52:1–121.
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30:2114–2120.
- Chen Z, Grover CE, Li P, Wang Y, Nie H, Zhao Y, Wang M, Liu F, Zhou Z, Wang X, et al. 2017. Molecular evolution of the plastid genome during diversification of the cotton genus. Mol Phylogenet Evol. 112:268–276.
- Hamzeh M, Dayanandan S. 2004. Phylogeny of Populus (Salicaceae) based on nucleotide sequences of chloroplast TRNT-TRNF region and nuclear rDNA. Am J Bot. 91:1398–1408.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Kim J, Kim Y, Park J. 2019. Complete chloroplast genome sequence of the Salix koriyanagi Kimura ex Goerz (Salicaceae). Mitochondrial DNA Part B. 4:549–550.
- Kim Y, Oh YJ, Han KY, Kim GH, Ko J, Park J. 2019. The complete chloroplast genome sequence of Hibicus syriacus L.‘Mamonde’ (Malvaceae). Mitochondrial DNA Part B. 4:558–559.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35:1547–1549.
- Kwon W, Kim Y, Park J. 2019. The complete chloroplast genome of Korean Marchantia polymorpha subsp. ruderalis Bischl. & Boisselier: low genetic diversity between Korea and Japan. Mitochondrial DNA Part B. 4:959–960.
- Lauron-Moreau A, Pitre FE, Argus GW, Labrecque M, Brouillet L. 2015. Phylogenetic relationships of American willows (Salix L., Salicaceae). PLoS One. 10:e0121965.
- Lee TB. 2003. Coloured flora of Korea. Seoul, Korea: Hayangmunsa.
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv preprint arXiv:13033997.
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The sequence alignment/map format and SAMtools. Bioinformatics. 25:2078–2079.
- Min J, Park J, Kim Y, Kwon W. 2019. The complete chloroplast genome of Artemisia fukudo Makino (Asteraceae): providing insight of intraspecies variations. https://www.tandfonline.com/doi/full/10.1080/23802359.2019.1601044
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32:268–274.
- Park J, Kim Y, Xi H, Kwon W, Kwon M. 2019. The complete chloroplast and mitochondrial genomes of Hyunsasi tree, Populus alba x Populus glandulosa (Salicaceae). DOI:10.1080/23802359.2019.1598788
- Shen X, Wu M, Liao B, Liu Z, Bai R, Xiao S, Li X, Zhang B, Xu J, Chen S. 2017. Complete chloroplast genome sequence and phylogenetic analysis of the medicinal plant Artemisia annua. Molecules. 22:1330.
- Wu J, Nyman T, Wang DC, Argus GW, Yang YP, Chen JH. 2015. Phylogeny of Salix subgenus Salix sl (Salicaceae): delimitation, biogeography, and reticulate evolution. BMC Evol Biol. 15:31.
- Wu Z. 2015. The new completed genome of purple willow (Salix purpurea) and conserved chloroplast genome structure of Salicaceae. J Nat Sci. 1:e49.
- Xia Y, Hu Z, Li X, Wang P, Zhang X, Li Q, Lu C. 2016. The complete chloroplast genome sequence of Chrysanthemum indicum. Mitochondrial DNA Part A. 27:4668–4669.
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18:821–829.
- Zhao QY, Wang Y, Kong YM, Luo D, Li X, Hao P. 2011. Optimizing de novo transcriptome assembly from short-read RNA-Seq data: a comparative study. BMC Bioinformatics. 12:S2.
