Abstract
The complete mitogenome sequence of the northern red-backed vole (Myodes rutilus) was determined using long polymerase chain reaction (PCR). The genome was 16,296 bp in length and contained 13 protein-coding genes, 2 ribosomal RNA genes, 22 transfer RNA genes, 1 origin of L strand replication, and 1 control region. The overall base composition of the heavy strand is A (32.9%), C (27.0%), T (26.9%), and G (13.2%). The base compositions present clearly the A–T skew, which is most obvious in the control region and protein-coding genes. Mitochondrial genome analyses based on MP, ML, NJ, and Bayesian analyses yielded identical phylogenetic trees. Results of phylogenetic analysis showed that Myodes had close relationship with Eothenomys, and had distant relationship with Microtus, Cricetulus, Dicrostonyx, Peromyscus, and other genera of Cricetidae. This study verifies the evolutionary status of M. rutilus in Myodes at the molecular level.
In this article, the complete mitochondrial genome of the northern red-backed vole (Myodes rutilus) was sequenced for the first time on ABI 3730XL using a primer walking strategy and the long and accurate polymerase chain reaction (PCR), with five pairs of long PCR primers and with 14 pairs of sub-PCR primers. A muscle sample was obtained from a female the northern red-backed vole captured from Mudanjiang region of Changbaishan Mountains in Heilongjiang Province, China (44°47′48″N, 129°04′52″ E). The specimen is stored in Animal and Plant Herbarium of Mudanjiang Normal University. The voucher number is MD2018044.
The mitochondrial genome is a circular double-stranded DNA sequence that is 16,296 bp long including 13 protein-coding genes, 2 rRNA genes, 22 tRNA genes, 1 origin of L strand replication, and 1 control region. The accurate annotated mitochondrial genome sequence was submitted to GenBank with accession number MK482363. The arrangement of the multiple genes is in line with other Cricetidae species (Triant and DeWoody Citation2006; Fan et al. Citation2011; Hao et al. Citation2011; Bendová et al. Citation2016; Chen et al. Citation2016; Cong et al. Citation2016; Kang et al. Citation2016; Luo and Liao Citation2016; Park et al. Citation2017) and most mammals (Meganathan et al. Citation2012; Yoon et al. Citation2013; Xu et al. Citation2012, Citation2013; Liu, Tian, Jin, Jin, et al. Citation2017; Liu, Tian, Jin, Dong, et al. Citation2017; Liu, Wang, et al. Citation2017; Liu et al. Citation2018; Liu, Dang, et al. Citation2019; Liu, Qin, et al. Citation2019; Jin et al. Citation2017).
The control region of the northern red-backed vole mitochondrial genome was located between the tRNA-Pro and tRNA-Phe genes, and contains only promoters and regulatory sequences for replication and transcription, but no structural genes. Three domains were defined in the northern red-backed vole mitochondrial genome control region (Zhang et al. Citation2009): the extended termination-associated sequence (ETAS) domain, the central conserved domain (CD), and the conserved sequence block (CSB) domain.
The total length of the protein-coding gene sequences was 11,392 bp. Most protein-coding genes initiate with ATG except for ND1, ND2, ND3, and ND5, which began with ATA, ATT, or GTG. Nine protein-coding genes terminated with TAA. The incomplete stop codons (T– – or TA–) were used in ND1, ATP6, COX3, and ND4. A strong bias against A at the third codon position was observed in the protein-coding genes. The frequencies of CTA (Leu), ATT (Ile), TTA (Leu), and ATA (Met) were higher than those of other codons. The length of tRNA genes varied from 59 to 75 bp. Twenty-one of them could be folded into the typical cloverleaf secondary structure except the tRNA-Ser(AGY), whose complete dihydrouridine arm was lacking.
Most of the northern red-backed vole mitochondrial genes were encoded on the H strand, except for the ND6 gene and eight tRNA genes, which were encoded on the L strand. Some reading frame intervals and overlaps were found. One of the most typical overlaping was between ATP8 and ATP6. The L-strand replication origin (OL) was located within the WANCY region containing five tRNA genes (tRNATrp, tRNA-Ala, tRNA-Asn, tRNA-Cys, and tRNA-Tyr). This region was 31 bp long and had the potential to fold into a stable stem-loop secondary structure. The total base composition of the northern red-backed vole mitochondrial genome was A (32.9%), C (27.0%), T (26.9%), and G (13.2%). The base compositions clearly present the A–T skew, which was most obvious in the control region and protein-coding genes.
In order to explore the evolution of Cricetidae species which include 28 genera, especially the evolution of genus Myodes, here, we investigate the molecular phylogenetics of the Chinese northern red-backed vole using complete mitochondrial genome sequence of 54 species. All sequences generated in this study have been deposited in the GenBank ().
Figure 1. Phylogenetic tree generated using the maximum parsimony method based on complete mitochondrial genomes. Akodon montensis (KF769456), Allocricetulus eversmanni (KP231506), Cricetulus kamensis (KJ680375), C. griseus (DQ390542), C. migratorius (KT918407), C. longicaudatus (KM067270), Cricetus cricetus (MF405145), Dicrostonyx torquatus (KX066190), D. groenlandicus (KX712239), D. hudsonius (KX683880), Eothenomys miletus (KX014874), E. chinensis (FJ483847), E. regulus (JN629046), E. inez (KU200225), Habromys ixtlani (KY707304), Isthmomys pirrensis (KY707312), Lasiopodomys mandarinus (KF819832), Meriones meridianus (KR013227), M. libycus (KR013226), M. tamariscinus (KT834971), Mesocricetus auratus (EU660218), Microtus rossiaemeridionalis (DQ015676), M. fortis (JF261175), M. levis (NC 008064), M. kikuchii (AF348082), M. ochrogaster (KT166982), M. arvalis (MG948434), M. agrestis (MH152570), Myodes glareolus (KF918859), M. rufocanus (KT725595), Neodon irene (HQ416908), N. sikimensis (KU891252), Neotoma mexicana (KY707300), N. magister (MG182016), Neotomodon alstoni (KY707310), Onychomys leucogaster (KU168563), Oligoryzomys stramineus (MF696155), Ondatra zibethicus (KX377613), Peromyscus maniculatus (MH260579), P. leucopus (MH256659), P. megalops (KY707305), P. crinitus (KY707308), P. melanophrys (KY707303), P. polionotus (KY707301), P. pectoralis (KY707309), P. aztecus (KY707306), P. attwateri (KY707299), Phodopus roborovskii (KU885975), Proedromys. liangshanensis (FJ463038), Podomys floridanus (KY707302), Reithrodontomys mexicanus (KY707307), Sigmodon hispidus (KY707311), Tscherskia triton (EU031048), and Wiedomys cerradensis (KF769457). The out group is Apodemus peninsulae (JN546584).
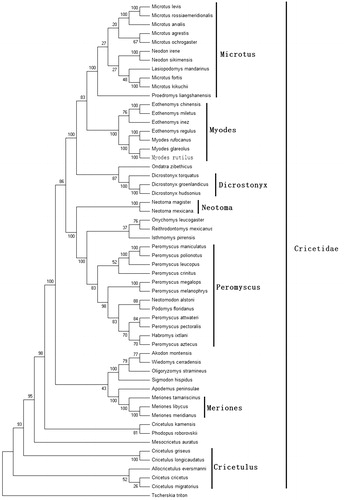
Mitochondrial genome analyses based on MP, ML, NJ, and Bayesian analyses yielded identical phylogenetic trees, indicating a close phylogenetic affinity of species. The phylogram obtained from maximum parsimony method is shown in . Results of phylogenetic analysis showed that Myodes had close relationship with Eothenomys, and had distant relationship with Microtus, Cricetulus, Dicrostonyx, Peromyscus, and other genera. This study verifies the evolutionary status of M. rutilus in Myodes at the molecular level. The mitochondrial genome would be a significant supplement for the M. rutilus genetic background.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the article.
Additional information
Funding
References
- Bendová K, Marková S, Searle JB, Kotlík P. 2016. The complete mitochondrial genome of the bank vole Clethrionomys glareolus (Rodentia: Arvicolinae). Mitochondrial DNA Part A. 27:111–112.
- Chen S, Chen G, Wei H, Wang Q. 2016. Complete mitochondrial genome of the Père David’s vole, Eothenomys melanogaster (Rodentia: Arvicolinae). Mitochondrial DNA Part A. 27:2496–2497.
- Cong H, Kong L, Liu Z, Wu Y, Motokawa M, Harada M, Li Y. 2016. Complete mitochondrial genome of the mandarin vole Lasiopodomys mandarinus (Rodentia: Cricetidae). Mitochondrial DNA Part A. 27:760–761.
- Fan L, Fan Z, Yue H, Zhang X, Liu Y, Sun Z, Liu S, Yue B. 2011. Complete mitochondrial genome sequence of the Chinese scrub vole (Neodon irene). Mitochondrial DNA. 22:50–52.
- Hao H, Liu S, Zhang X, Chen W, Song Z, Peng H, Liu Y, Yue B. 2011. Complete mitochondrial genome of a new vole Proedromys liangshanensis (Rodentia: Cricetidae) and phylogenetic analysis with related species: are there implications for the validity of the genus Proedromys? Mitochondrial DNA. 22:28–34.
- Jin ZM, Liu Z, Ma JZ. 2017. Sequencing and analysis of the complete mitochondrial genome of the masked shrew (Sorex caecutiens) from China. Mitochondrial DNA Part B. 2:486–488.
- Kang C, Yue H, Liu M, Huang T, Liu Y, Zhang X, Yue B, Zeng T, Liu S. 2016. The complete mitochondrial genome of Cricetulus kamensis (Rodentia: Cricetidae). Mitochondrial DNA Part A. 27:976–977.
- Liu Z, Bai W, Wang AN, Tian XM, Li DW. 2018. Sequencing and analysis of the complete mitochondrial genome of the taiga shrew (Sorex isodon) from China. Mitochondrial DNA Part B. 3:466–468.
- Liu Z, Dang YQ, Li JJ. 2019. Sequencing and analysis of the complete mitochondrial genome of the Eurasian least shrew (Sorex minutissimus) from China. Mitochondrial DNA Part B. 4:178–180.
- Liu Z, Qin KS, Li JJ, Dong M. 2019. Sequencing and analysis of the complete mitochondrial genome of the Siberian large-toothed shrew (Sorex daphaenodon) from China. Mitochondrial DNA Part B. 4:542–544.
- Liu Z, Tian XM, Jin JL, Jin ZM, Li DW, Zhang JS. 2017. Sequencing and analysis of the complete mitochondrial genome of the slender shrew (Sorex gracillimus) from China. Mitochondrial DNA Part B. 2:642–644.
- Liu Z, Tian XM, Jin ZM, Dong M, Zhang JS. 2017. Sequencing and analysis of the complete mitochondrial genome of the Ussuri shrew (Sorex mirabilis) from China. Mitochondrial DNA Part B. 2:645–647.
- Liu Z, Wang AN, Zhang JS, Yang X, Liu H. 2017. Sequencing and analysis of the complete mitochondrial genome of flat-skulled shrew (Sorex roboratus) from China. Mitochondrial DNA Part B. 2:369–371.
- Luo G, Liao J. 2016. The complete mitochondrial genome of Allocricetulus eversmanni (Rodentia: Cricetidae). Mitochondrial DNA Part A. 27:3102–3104.
- Meganathan PR, Pagan HJT, McCulloch ES, Stevens RD, Ray DA. 2012. Complete mitochondrial genome sequences of three bats species and whole genome mitochondrial analyses reveal patterns of codon bias and lend support to a basal split in Chiroptera. Gene. 492:121–129.
- Park YJ, Park CE, Hong SJ, Jung BK, Ibal JC, Park GS, Shin JH. 2017. The complete mitochondrial genome sequence of the greater wax moth Galleria mellonella (Insecta, Lepidoptera, Pyralidae): sequence and phylogenetic analysis comparison based on whole mitogenome. Mitochondrial DNA Part B. 2:714–715.
- Triant DA, DeWoody JA. 2006. Accelerated molecular evolution in Microtus (Rodentia) as assessed via complete mitochondrial genome sequences. Genetica. 128:95–108.
- Xu CZ, Zhang HH, Ma JZ. 2013. The complete mitochondrial genome of sable, Martes flavigula. Mitochondrial DNA. 24:240–242.
- Xu CZ, Zhang HH, Ma JZ, Liu ZH. 2012. The complete mitochondrial genome of sable, Martes zibellina. Mitochondrial DNA. 23:167–169.
- Yoon KB, Kim HR, Kim JY, Jeon SH, Park YC. 2013. The complete mitochondrial genome of the Ussurian tube-nosed bat Murina ussuriensis (Chiroptera: Vespertilionidae) in Korea. Mitochondrial DNA. 24:397–399.
- Zhang HH, Xu CZ, Ma JZ. 2009. Structure of the mtDNA control region and phylogeny of the Mustelidae species. Acta Ecol Sin. 29:3585–3592.
