Abstract
Gambian rats are found in central Africa, in regions south of the Sahara desert as far south as Zululand. Herein, we describe the first rat assembly mtDNA in Cricetomys genera, which contains 13 protein-coding genes (PCGs), two rRNA genes (12S rRNA and 16S rRNA), 22 transfer RNA (tRNA) genes, and a control region (D-loop). The complete mitochondrial genome sequence provided here would be useful for further understanding the evolution of Cricetomys and conservation genetics of Cricetomys gambianus.
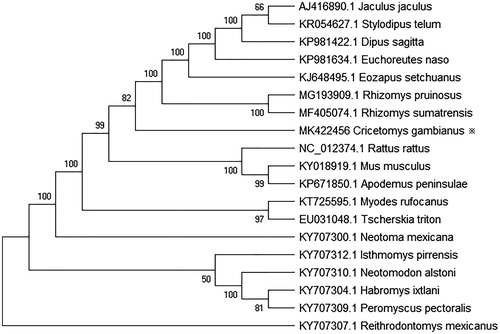
Gambian rats have coarse, brown fur and a dark ring around the eyes (Nowak and Walker Citation1997). Their long tails are scaly and they have narrow heads with small eyes. The main physical characteristic of Gambian rats and all Cricetomys, in general, are their large cheek pouches (Ryan Citation1989). These pouches can expand to a great size, allowing Gambian rats to transport massive quantities of food if necessary. Cheek pouches also exist in other families of rodentia, such as the African hamster and members of the subfamily Cricetinae. Males and females are usually the same size, with little sexual dimorphism. Gambian rats can reach sizes up to 910 mm and beyond, including the tail. These rats also have a very low fat content, which may be the cause of their susceptibility to cold (Bobe and Mabela Citation1997). No mitochondrial genome of Cricetomys gambianus was available until now. We will determine the mitochondrial genome of C. gambianus in this study. The sample of C. gambianus was obtained in Florida, USA. And its DNA was extracted from the liver of an adult female C. gambianus, then sequenced with the TruSeq DNA PCR-Free Library Preparation Kit (Illumina), as recommended. Data was stored as well as submitted by the National Institute of Environmental Health Sciences (NIEHS), the sample of Sistrurus miliarius was stored in NCBI (Accession no. SAMN03754047). The complete mitogenome was assembled using MIRA 4.0.2 (San Francisco, CA) (Burlibasa and Vasiliu Citation1999) and MITObim 1.9 (Hahn et al. Citation2013) and using Rattus rattus (NC_012374.1) as the reference sequence. And then, the mitochodrial genome was annotated and drawn with MitoFish 3.30 (http://mitofish.aori.u-tokyo.ac.jp/). The complete mitochondrial genome of C. gambianus is a double-stranded, circular DNA 16,559 bp in total length (GenBank accession no. MK422456), and includes 13 protein-coding genes, 2 ribosomal RNA genes (12S rRNA and 16S rRNA), 22 tRNA genes, and a control region (D-loop). The contents of A, T, G, and C are 35.74%, 27.62%, 11.32%, and 25.32%, respectively. GC contents of mitochondrial genome are 36.64%. Thirteen of the PCGs use complete (ATG, ATT, ATA) start codon and nine of PCGs have complete stop codon(TAA, TAG), which also proves the integrity of this assembly. The 12S rRNA and 16S rRNA genes are 952 and 561 bp long, respectively. The lengths of 22 tRNA genes range from 58 bp (tRNA-Ser) to 75 bp (tRNA-Leu). The D-loop length is 1151 bp and lies between the tRNA-Phe and antisense tRNA-Pro genes. Phylogenetic analysis of 20 mitochondrial genomes using MEGA 7 (Kumar et al. Citation2016) using Reithrodontomys mexicanus as an outer population indicated that C. gambianus and Rhizomys sumatrensis are the most closely related species (). The mitogen of Sistrurus miliarius helps to understand the phylogeny and evolution of the rat.
Figure 1. Neighbor-joining molecular phylogenetic tree of 19 species based on complete mitogenome sequences, with R. mexicanus as outgroup. The asterisk indicates the individual sampled in this study.
As a conclusion, we obtained and described the complete mitotic genome from C. gambianus, which constitutes a valuable and useful resource for population genetic research and monitoring, as well as further protection of the species.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Bobe L, Mabela M. 1997. Incidence of four gastro-intestinal parasite worms in group of cricetomas, Cricetomys gambianus (Rodent: Cricetidae), caught at Lukaya-Democratic Republic of Congo. Tropicultura. 15:132–135.
- Burlibasa CVD, Vasiliu M. 1999. Genome sequence assembly using trace signals and additional sequence information. German Conf Bioinform. 99:45–56.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads–a baiting and iterative mapping approach. Nucleic Acids Res. 41:e129.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- Nowak RM, Walker EP. (1997). Mammals of the world. Baltimore, MD: John Hopkins University Press.
- Ryan J. 1989. Evolution of cheek pouches in African pouched rats (Rodentia: Cricetomyinae). J Mammal. 70:267–274.
