Abstract
In this study, the complete mitochondrial DNA sequence was obtained from Dactylellina leptospora using next-generation sequencing technology. The mitogenome is a circular molecule of 201,677 bp in length. The overall nucleotide composition was 38.54% A, 36.34% T, 11.21% C, and 13.9% G, with a higher A + T content (74.88%) than other species. In the D. leptospora mitogenome, we identified 36 genes including 15 protein-coding genes, 2 ribosomal RNA (rRNA), 19 transfer RNA (tRNA). Phylogenetic analysis showed D. leptospora was genetically divergent from species Pyronema omphalodes within Pezizomycetes.
Dactylellina leptospora, which belongs to order Orbiliales, family Orbiliaceae, is a nematode-trapping fungus. D. leptospora reported as the type species of both Dactylosporium and Dactylellina (Rubner Citation1996). Also, these species trap the nematodes by stalked knobs and non-constricting (Drechsler Citation1937). Here, we determined the complete mitochondrial genome sequence of D. leptospora and its phylogenetic relationships with other related species based on concatenated amino acid sequences of 15 mitochondrial protein-coding genes for the first time.
The strain of D.leptospora sequenced in this study was isolated from soil (at Dali Bai Autonomous Prefecture, Yunnan province, by Jing Zhang, 2004. N24°41'E 98°52') and has been deposited and stored in the formerly Key Laboratory of Industrial Microbiology and Fermentation Technology of Yunnan (University of Yunnan in Kunming, China). Mitogenome assembly was performed by the software SPAdes 3.9.0 (Bankevich et al. Citation2012). The mitogenome was annotated using MFANNOT and was manually adjusted.
The complete mtDNA sequence of D. leptospora reported here has been deposited in GenBank under the accession number MK307795. The complete mitochondrial genome of D. leptospora was 201,677 bp in size and its overall base composition is 38.54% for A, 11.21% for C, 13.9% for G, and 36.34% for T. It contains 36 genes, including ribosomal protein S3 (rps3), 3 subunits of ATPase (atp6, atp8, and atp9), 3 subunits of cytochrome c oxidase (cox1, cox2, and cox3), 7 subunits of NADH dehydrogenase (nad1, nad2, nad3, nad4, nad4L, nad5, and nad6) and apocytochrome b (cob), and 19 tRNA(tRNAGln, tRNAAla, tRNAAsn, 3tRNAArg, tRNALys, tRNAGly, tRNAVal, tRNAThr, tRNAHis, 2tRNASer,tRNATyr, tRNAPhe, tRNAGlu, and 3tRNAMet) and 2 rRNA. All the predicted genes totally possessed 58 introns, but none of the tRNAs possessed introns.
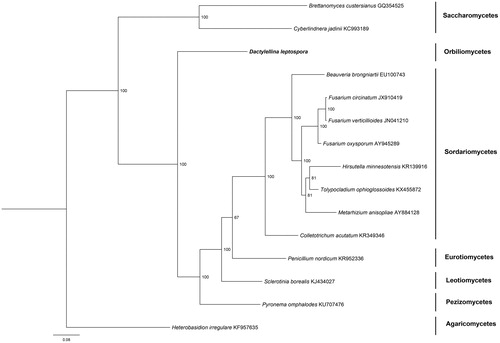
The phylogenetic tree among 15 ascomycetes species based on 15 protein-coding genes was visualized using Mrbayes (Ronquist and Huelsenbeck Citation2003). As shown in . Phylogenetic analysis confirmed the placement of D. leptospora, as it is clearly divergent from Pyronema omphalodes.
Figure 1. The phylogenetic tree of the Dactylellina leptospora mitogenome and 14 related organisms based on 15 conserved mitochondrial protein-coding genes (atp6, atp8, atp9, cytb, cox1, cox2, cox3, nad1, nad2, nad3, nad4, nad4L, nad5, nad6, and rps3). The 15 conserved mitochondrial protein-coding genesis downloaded from GenBank and the phylogenic tree was generated using Bayesian inference (BI). The percentages of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000,000 replicates) were shown next to the branches.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19:455–477.
- Drechsler C. 1937. Some hyphomycetes that prey on free-living terricolous nematodes. Mycologia. 29:447–552.
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 19:1572–1574.
- Rubner A. 1996. Revision of predacious hyphomycetes in the Dactylella-Monacrosporium complex. Stud Mycol. 39:1–134.
