Abstract
Ottelia acuminata var. songmingensis is an Endangered freshwater submerged macrophyte occupying only a portion of a stream ca. 300 meters long in Songming County of the Yunnan Province, China. Here, we report and characterize the complete chloroplast genome sequence of O. acuminata var. songmingensis to provide genomic resources useful for promoting its conservation. The complete chloroplast genome is 157,645 bp in length, consisting of a pair of inverted repeat (IR) regions (24,973 bp), a small single copy (SSC) region (20,055 bp), and a large single copy (LSC) region (87,644 bp). The chloroplast genome contained 116 genes, including 71 protein-coding genes, 37 tRNA genes, and 8 rRNA genes. The overall GC content of the whole genome is 36.6%. Phylogenetic analysis of 19 selected chloroplast genomes revealed that O. acuminata var. songmingensis was closely related to Elodea canadensis of Hydrocharitaceae.
Ottelia acuminata (Gagnepain) Dandy (Hydrocharitaceae) is an endemic submerged perennial in China (Wang et al. Citation2010). It has been categorized as vulnerable with a status ‘A2c’ in the China Species Red List (Qin et al. Citation2017). During the past 30 years, O. acuminata has disappeared from 12 lakes or rivers where it previously had been common (Guo et al. Citation2019). Unfortunately, the rapid disappearance of populations continues. Ottelia acuminata (Gagnepain) Dandy var. songmingensis Z. T. Jiang, H. Li et Z. L. Dao (Jiang et al. Citation2005) is a variant that only occurs in Songming County of the Yunnan Province occupying a portion of a stream ca. 300 meters long. Here, we reported the complete cp genome of O. acuminata var. songmingensis, to provide a genomic resource for further conservation genetics and phylogenetic analysis.
The fresh leaves of O. acuminata var. songmingensis were collected from Songming County (25°16′23.2″N, 102°52′52.7″E), Yunnan province and voucher specimens (Guo SM01) were deposited in Herbarium of Yunnan Normal University. A sequence library was constructed and sequencing was performed using the Illumina HiSeq 2500-PE150 platform (Illumina, San Diego, CA, USA). All raw reads were filtered by using NGS QC Toolkit_v2.3.3 with default parameters to obtain clean reads (Patel and Jain Citation2012). The plastome was de novo assembled using NOVOPlasty (Dierckxsens et al. Citation2017). The complete cp genome was annotated with the online annotation tool GeSeq (Tillich et al. Citation2017).
The complete chloroplast genome of O. acuminata var. songmingensis was a quadripartite circular and 157,645 bp in length (GenBank accession No.: MK604184), comprising a large single copy (LSC) of 87,644 bp and a small single copy (SSC) of 20,055 bp, separated by two inverted repeat (IR) regions of 24,973 bp. It contained 116 genes, including 71 protein-coding, 8 ribosomal RNA, and 37 tRNA genes. The base compositions of the chloroplast genome were uneven (31.2% A, 18.7% C,18.0%G, 32.1% T), with an overall GC content of 36.6%. A total of 79 SSRs were discovered by the online software IMEx (Mudunuri and Nagarajaram Citation2007). Among them, the numbers of mono-, di-, tri-, tetra-, penta- and hexa- nucleotides SSRs are 24, 33, 8, 11, 2, and 1, respectively.
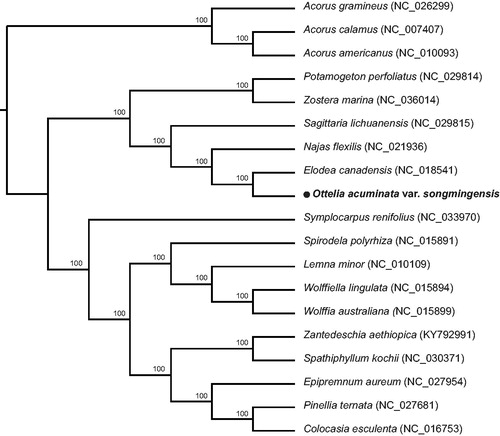
To determine the phylogenetic location of O. acuminata var. songmingensis, the published chloroplast genomes, including Elodea canadensis (NC_018541, Huotari and Korpelainen Citation2012) from Hydrocharitaceae and 14 species from Alismatales were used, with 3 Acorus species from Acorales as outgroups. The total of 19 chloroplast genomes were aligned using MAFFT 7.308 (Katoh and Standley Citation2013). The maximum likelihood (ML) tree was reconstructed by RAxML 8.2.11 (Stamatakis et al. Citation2008) with the nucleotide substitution model of GTR + G and node support was estimated by means of bootstrap analysis with 1000 replicates. The phylogenetic tree showed that O. acuminata var. songmingensis and Elodea canadensis from Hydrocharitaceae formed a monophyletic clade with 100% bootstrap value (). The complete chloroplast genome of O. acuminata var. songmingensis will provide a useful resource for the conservation genetics of this species as well as for the phylogenetic studies of Hydrocharitaceae.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:e18.
- Guo JL, Yu YH, Zhang JW, Li ZM, Zhang YH, Volis S. 2019. Conservation strategy for aquatic plants: Endangered Ottelia acuminata (Hydrocharitaceae) as a case study. Biodivers Conserv. 28:1533–1548.
- Huotari T, Korpelainen H. 2012. Complete chloroplast genome sequence of Elodea canadensis and comparative analyses with other monocot plastid genomes. Gene. 508:96–105.
- Jiang Z, Li H, Dao Z. 2005. Ottelia acuminata var. songmingensis, a new variety of the Hydrocharitaceae from Yunnan, China. Guihaia. 25:424–425.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Mudunuri SB, Nagarajaram HA. 2007. IMEx: Imperfect Microsatellite Extractor. Bioinformatics. 23:1181–1187.
- Patel RK, Jain M. 2012. NGS QC toolkit: a toolkit for quality control of next generation sequencing data. PLos One. 7:e30619.
- Qin H, Yang Y, Dong S, He Q, Jia Y, Zhao L, Yu S, Liu H, Liu B, Yan Y, et al. 2017. Threatened species list of China’s higher plants. Biodivers Sci. 25:696–744.
- Stamatakis A, Hoover P, Rougemont J. 2008. A rapid bootstrap algorithm for the RAxML web servers. Syst Biol. 57:758–771.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45:W6–W11.
- Wang QF, Guo YH, Haynes RR, Hellquist CB. 2010. Hydrocharitaceae. In: Wu ZY, Peter RH, Hong DY, editors. Flora of China. Vol. 23. St Louis: Science Press, Beijing and Missouri Botanical Garden Press; pp. 91–102.