Abstract
Rhus chinensis is an important economic species, which could provide raw materials for pharmaceutical and industrial dyes. This is the first report of R. chinensis chloroplast genomes by de novo sequencing. The results showed that the length of R. chinensis was 159,082 bp. The length of LSC and SSC was 85,394 bp and 18,663 bp, respectively. The genomes contained 126 genes, including 88 protein encoding genes, eight rRNA, and 30 tRNA genes. The clustering results showed that Anacardiaceae were closest to R. chinensis, followed by Aceraceae and Anacardiaceae.
Rhus chinensis are important tolerant species because they are resistant drought, cold, and saline-alkali conditions (Shim et al. Citation2003). Moreover, R. chinensis also has important economic value and medicinal value (Zhou et al. Citation2016). However, due to human activities and overexploitation, R. chinensis has been seriously damaged. The chloroplast genome plays an important role in the study of genetic evolution (Tian and Li Citation2002; Bremer et al. Citation2002; Leliaert et al. Citation2012). Up to now, there is no related research about the chloroplast genome of Rhus up till the present moment, which seriously hinder the molecular genetic research of Rhus. Here, we present the first complete chloroplast genomes of R. chinensis by de novo sequencing.
We collected the experimental materials (stored in herbarium of West Anhui University) from College of Biological and Pharmaceutical Engineering (N:31°45′55.94″, E:116°31′30.00″China). High salt and low pH method were used to extracted cp-DNA for high-throughput sequencing. The full-length sequence of chloroplast genome was 158,809 bp, with 85,394 bp length of LSC, 18,663 bp of SSC and 27,376 bp of two IR regions (uploaded to NCBI with the number of MG267385). The GC content was 37.86%. The chloroplast genome encoded 126 genes, including 88 protein-coding genes, eight rRNA, and 30 tRNA genes. In the chloroplast genome, about 48.80% of the sequence is the gene coding region.
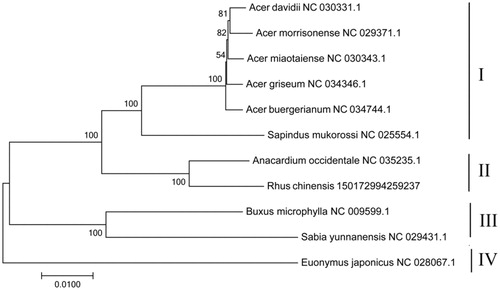
To determine the phylogenetic positions of R. chinensis in sapindales plants, we performed a phylogenetic analysis using 66 common chloroplast protein-coding genes from other 10 sapindales plants species published in NCBI. All 11 plants were divided into four categories. The first category consisted of six species that consist of five Aceraceae species and one Sapindaceae species, which showed that Aceraceae and Sapindaceae have close relationship. The second category was consisted by Anacardium occidentale and R. chinensis. Moreover, the third category was also consisted by two species: Buxus microphylla and Sabia yunnanensis; The Euonymus japonicas was far apart from other plants and is divided into one category. The clustering results showed that Anacardiaceae were closest to R. chinensis, followed by Aceraceae and Anacardiaceae. This study provides a molecular basis for the classification of R. chinensis.
shows the ML phylogenetic tree of the R. chinensis clade based on same protein-coding genes. Numbers above or below the nodes are bootstrap support values.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Bremer B, Bremer K, Heidari N, Erixon P, Olmstead RG, Anderberg AA, Källersjö M, Barkhordarian E. 2002. Phylogenetics of asterids based on 3 coding and 3 non-coding chloroplast DNA markers and the utility of non-coding DNA at higher taxonomic levels. Mol Phylogenet Evol. 24:274–301.
- Leliaert F, Smith DR, Moreau H, Herron MD, Verbruggen H, Delwiche CF, Clerck OD. 2012. Phylogeny and molecular evolution of the green algae. Crit Rev Plant Sci. 31:1–46.
- Shim YJ, Doo HK, Ahn SY, Kim YS, Seong JK, Park IS, Min BH. 2003. Inhibitory effect of aqueous extract from the gall of Rhus chinensis on alpha-glucosidase activity and postprandial blood glucose. J Ethnopharmacol. 85:283–287.
- Tian X, Li DZ. 2002. App lication of DNA sequences in plant phylogenetic study. Acta Botanica Yun-Nanica. 24:170–184.
- Zhou CF, Huang XY, Li Y, Luo JW, Cai LP. 2016. Changes in subcellular distribution and antioxidant compounds involved in Pb accumulation and detoxification in Neyraudia reynaudiana. Environ Sci Pollut Res. 23:21794–21804.