Abstract
The complete mitochondrial (mt) genome of Monolepta occifuvis Gressitt & Kimoto (Coleoptera: Chrysomelidae: Galerucinae) was sequenced with 15,998 bp long. The sequenced genome has the standard metazoan complement of 37 genes and an A + T-rich region which is consistent with insect ancestral genome arrangement. All protein-coding genes (PCGs) initiate with ATN except that nad1 initiates with TTG. The control region occupied 1,407 bp with two conserved blocks. The phylogenetic results support Alticinae as a separate subfamily and being the sister group to Galerucinae while the monophyly of Galerucinae was not recovered.
Monolepta occifuvis Gressitt & Kimoto belongs to Galerucinae (Coleoptera: Chrysomelidae), which is one of the largest assemblages of leaf beetles. Five tribes system in Galerucinae was proposed which is very stable till now (Senno and Wilcox Citation1982). However, whether Alticinae should be a separate subfamily as the sister group to Galerucinae among Chrysomelidae (Furth and Suzuki Citation1998) or to be a tribe within Galerucinae (Duckett et al. Citation2004) is unresolved. Monolepta occifuvis is one of the most important pests for Euphoria longan (Lour.) Steud., whose adults endangers the young shoots. They cause mass reductions and serious economic losses. However, there is no mitochondrial (mt) genome sequence of M. occifuvis was published to date.
Here, we determined the first complete mt genome of M. occifuvis (GenBank accession number: MK409736) containing 37 genes (13 PCGs, 22 tRNA genes, and two rRNA genes) typically being present in metazoan mt genomes (Wolstenholme Citation1992). The specimen used was collected by Juan Yang at 2017.10.28 in the Orchard of Guangxi University and the voucher specimens are deposited in the Entomological Museum of Guangxi University, Beijing. The 13 PCGs exhibit the canonical mt start codons for invertebrate mtDNAs (Wolstenholme Citation1992) with TTG for nad1 and ATN for the remaining 12 PCGs. As for stop codons, TAA, TAG, TGA as well as T-tRNA were used. The entire complement of 22 typical tRNAs in the arthropod mt genomes was found in M. occifuvis ranging from 55 to 71 bp. The rrnL is 1,275 bp long while the rrnS is 744 bp long.
Gene overlaps were found at eight gene junctions and involved a total of 23 bp; the longest overlap (8 bp) existed between tRNATrp and tRNACys as well as tRNATyr and COX1. There is a major non-coding region (control region) between the srRNA and tRNAIle occupying 1,407 bp, which includes two conserved blocks: one consisting of four 41 bp repeated units (AAGCTCAAATGTACGTAAATCCAATATACAATGAGTTTCCC) and the other consisting of three 10 bp repeated units (AAAAAAATGC). In addition to the large non-coding region, several small non-coding intergenic spacers are present in the M. occifuvis mt genome and spread over 16 positions, ranging in size from 1 to 19 bp.
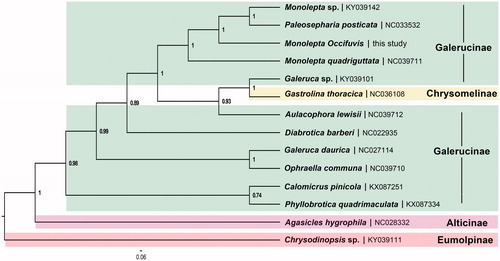
Phylogenetic tree was constructed with MrBayes 3.2.2 based on the concatenation data of 13 PCGs (only containing the first and the second codon positions) from 11 species of Galerucinae (). Chrysomelinae, Alticinae, and Eumolpinae were used as outgroups. The Alticinae was recovered as the sister group to Galerucinae, which support Alticinae as a separate subfamily. The monophyly of Galerucinae was not recovered as Gastrolina thoracica (Chrysomelinae) with in it, which need more comprehensive samples from Chrysomelidae to further study.
Disclosure statement
The authors report no conflict of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Duckett C, Gillespie J, Kjer K. 2004. Relationships among the subfamilies of Chrysomelidae inferred from small subunit ribosomal DNA and morphology, with special emphsis on the relationship among the flea beetles and the Galerucinae. In: Jolivet PH, Santiago-Baly JA, Schmitt M (eds), New developments in the biology of Chrysomelidae. Boston: Kluwer Academic publishers. 3–18.
- Furth DG, Suzuki K. 1998. Studies of Oriental and Australian Alticinae genera based on the comparative morphology of the metafemoral spring, genitalia, and hind wing venation. In: Furth DG, editor, Proceedings of the Fourth International Symposium on the Chrysomelidae. Museo Regionale di Scienze Naturali; p. 99–124.
- Senno T, Wilcox J. 1982. Leaf beetle genera (Coleoptera: Chrysomelidae). Entomography. 1:1–222.
- Wolstenholme DR. 1992. Animal mitochondrial DNA: structure and evolution. Int Rev Cytol. 141:173–216.