Abstract
The black-bellied hornet (Vespa basalis) is one of the most dangerous social wasps. In this study, the complete mitogenome sequence of V. basalis (Hymenoptera, Vespidae) has been sequenced (GenBank accession number MK440075). The mitogenome, 16,735 bp, includes 13 protein-coding, 22 transfer RNAs, two ribosomal RNAs genes, and a noncoding D-loop region. The D-loop of 807 bp length is located between rRNA-S and tRNATyr. The overall base composition of V. basalis is 41.82% for A, 12.09% for C, 40.53% for T, and 5.57% for G, with a high AT bias of 82.35%. The present data could contribute to detailed phylogeographic analysis of this dangerous wasp.
Vespa basalis (Hymenoptera, Vespidae) is a species of social wasp, mainly distributed in China, Nepal, India, Thailand, Myanmar, Sikkim, Vietnam, Sri Lanka, and Indonesia. It said that this species is one of the most aggressive wasps, and the toxicity of its venom is strong (Ho and Ko Citation1988). However, this wasp is popular as food in Southwest China. Elucidating the structure of V. basalis mitogenome is important for understanding its diversity and evolution.
The specimen of V. basalis was obtained from Luquan County, Kunming, Yunnan, China (N 25°55′, E 102°.47′), and deposited in the Insect Collection of Research Institute of Resource Insects with an accession number RIRI-w-20181026. Sequencing work of the complete mitogenome of V. basalis was performed using Illumina Nextseq500 in Beijing Microread Genetics Co., Ltd., with a total data volume 4G (150 bp Reads), and high-quality reads were assembled from scratch using IDBA-UD and SPAdes (Gurevich et al. Citation2013). Protein-coding genes (PCGs) of the V. basalis mitogenome were identified using BLAST search in NCBI, and tRNA genes were identified using the tRNAscan-SE search server (Schattner et al. Citation2005).
The gene order and orientation of V. basalis mitogenome are identical to the most common type suggested as ancestral for insects (Boore et al. Citation1998; Taanman Citation1999). It was 16,735 bp in size (GenBank accession number MK440075), including 13 typical invertebrate PCGs, 22 transfer RNA genes, two ribosomal RNA genes, and a noncoding control region (D-loop). The A + T content of the whole V. basalis mitogenome is 82.35%, showing an obvious AT mutation bias (Eyre-Walker Citation1997). The D-loop region exhibits the highest A + T content (88.12%) in the V. basalis mitogenome. This region has been shown to harbor the origin sites for the transcription and replication for both strands of insect mitogenomes (Yukuhiro et al. Citation2002).
All PCGs use standard ATN as a start codon. As for the stop codon, 12 PCGs had the common stop codon TAA while COX3 terminated with incomplete stop codon T. Similar cases could be found in other insect mitogenomes (Yin et al. Citation2012). It has been demonstrated that the incomplete stop codons could produce functional stop codons “TAA” in polycistronic transcription cleavage and polyadenylation processes by the addition of 3’A residues (Ojala et al. Citation1981).
Like other wasp mitogenome, the rRNA-S gene in V. basalis mitogenome is located between tRNAVal and the D-loop region while rRNA-L is located between ND1 and tRNAVal (Zhao et al. Citation2019). All the tRNAs except tRNASe (AGN) could be folded into the typical cloverleaf secondary structures. The unusual tRNASe (AGN) lacks dihydrouridine (DHU) arm.
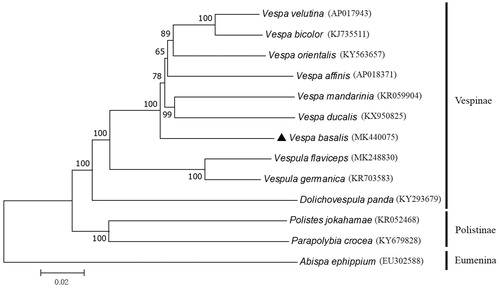
Based on the concatenated 13 PCGs sequences of 13 wasps mitogenomes, the neighbor-joining method was used to construct the phylogenetic relationship of V. basalis with 12 other wasps (). The result showed that V. basalis was the earliest independent branch within the genus Vespa (Hymenoptera, Vespidae), adding new molecular evidence into evolutionary history data of the Vespidae (Lopez-Osorio et al. Citation2014). This mitogenome data might be also useful for further phylogeography analyses in Hymenoptera.
Disclosure statement
The authors declare no competing materials in the preparation and execution of this manuscript. The authors are responsible for the content and writing of this paper.
Additional information
Funding
References
- Boore JL, Lavrov DV, Brown WM. 1998. Gene translocation links insects and crustaceans. Nature. 392:667–668.
- Ho CL, Ko JL. 1988. Purification and characterization of a lethal protein with phospholipase A1 activity from the hornet (Vespa basalis) venom. Biochimica et Biophysica Acta. 963:414–422.
- Eyre-Walker A. 1997. Differentiating between selection and mutation bias. Genetics. 147:1983–1987.
- Gurevich A, Saveliev V, Vyahhi N, Tesler G. 2013. QUAST: quality assessment tool for genome assemblies. Bioinformatics. 29:1072–1075.
- Lopez-Osorio F, Pickett KM, Carpenter JM, Ballif BA, Agnarsson I. 2014. Phylogenetic relationships of yellowjackets inferred from nine loci (Hymenoptera: Vespidae, Vespinae, Vespula and Dolichovespula). Mol Phylogenet Evol. 73:190–201.
- Ojala D, Montoya J, Attardi G. 1981. tRNA punctuation model of RNA processing in human mitochondria. Nature. 290:470–474.
- Schattner P, Brooks AN, Lowe TM. 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 33:W686.
- Taanman JW. 1999. The mitochondrial genome: structure, transcription, translation and replication. Biochim Biophys Acta. 1410:103–123.
- Yin H, Zhi Y, Jiang H, Wang P, Yin X, Zhang D. 2012. The complete mitochondrial genome of Gomphocerus tibetanus Uvarov, 1935 (Orthoptera: Acrididae: Gomphocerinae). Gene. 494:214–218.
- Yukuhiro K, Sezutsu H, Itoh M, Shimizu K, Banno Y. 2002. Significant levels of sequence divergence and gene rearrangements have occurred between the mitochondrial genomes of the wild mulberry silkmoth, Bombyx mandarina, and its close relative, the domesticated silkmoth, Bombyx mori. Mol Biol Evol. 19:1385–1389.
- Zhao M, Wang CY, Wang JD, He Z, Xu HC, Feng Y. 2019. The complete mitochondrial genome of an edible wasp Vespula flaviceps (Hymenoptera, Vespidae). Mitochondrial DNA Part B. 4:1085–1086.