Abstract
In the present study, the complete sequence of the mitochondrial genome (mitogenome) of Parasa tessellata (Lepidoptera: Limacodidae) is described. The mitogenome (15,308 bp) of P. tessellata encodes 13 protein-coding genes (PCGs), 22 transfer RNA genes (tRNAs), two ribosomal RNA genes (rRNAs), and an adenine (A) + thymine (T)-rich region. Its gene complement and order are similar to that of other sequenced lepidopterans. Phylogenetic analyses based on 13 PCGs using maximum likelihood (ML) and Bayesian inference (BI) revealed that P. tessellata resides in the Sphingidae family. This study provided the valuable evidence on phylogenetic relationship of the P. tessellata at the molecular level and essential resource for further research on this species.
Parasa tessellata (Lepidoptera: Limacodidae) is an endemic species to China which is distributed in Zhejiang, Jiangxi and Shanxi province. The species has largely infected forest plant populations; as well as it damages the roadside and garden trees around urban areas. Parasa tessellata distributed in low-altitude mountain areas. The larvae of this species are generally scorpion-like, have no gastropods, and slide with a suction cup. The back of the body is usually densely burr-like, toxic, and can cause skin rashes. Till now, there was less information about the complete mitochondrial genomes of P. tessellata in GenBank. In our study, the complete mitogenome of P. tessellata was determined, which would be useful for further genetic studies, phylogenetic analysis, and conservation of this species.
The sample of P. tessellata was obtained at Hebei village (27°42′13.8″N 115°41′16.6″E) in Jiangxi, China. The collected individuals were identified as P. tessellata by the taxonomist (Department of Entomology, AHAU) carefully observed the morphological characteristics and referenced Chinese moths published by Institute of Zoology, Chinese Academy of Sciences to ensure we identified species correctly, then preserved in 100% ethanol and stored at −80 °C. The total DNA was extracted using the Genomic DNA Extraction Kit (Aidlab Co., Beijing, China) to the manufacturer instructions. The sequence was amplified using PCR with twelve pairs of primer.
In general, the length of this sequence is 15,308 bp (Genbank No: MK599146), containing two ribosomal RNA genes, 22 putative transfer RNA (tRNA) genes, 13 protein-coding genes, and an adenine (A) + thymine (T)-rich region, consisted with previous studies (Sun et al. Citation2016). The 12 PCGs initiated by ATN codons except for cytochrome c oxidase subunit 1 (cox1) gene that is seemingly initiated by the CGA codon as documented in other insect mitogenomes (Dai et al. Citation2015; Sun et al. Citation2017). The 22 tRNA were scattered among the whole mitochondrion, and they range from 63 (tRNASer) to 74 (tRNALeu) in length. The A + T-rich region is 365 bp long, and contains some conserved regions, including ‘ATAGA’ motif followed by a 19 bp poly-T stretch, a microsatellite-like element (AT)7 and a poly-A element. The 12S rRNA (797bp) and 16S rRNA (1357bp) were separated by tRNAVal (Wu et al. Citation2013).
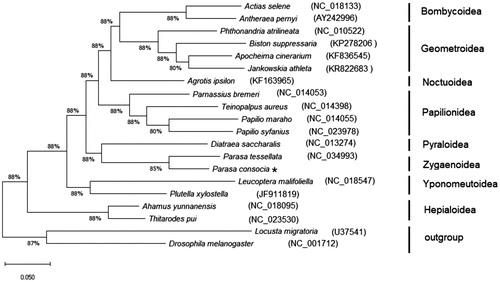
To reconstruct the phylogenetic relationship among Lepidopterans, 19 complete or near-complete mitogenomes were downloaded from the GenBank database. We reconstructed the phylogenetic relationships using ML method based on concatenated nucleotide sequences of the 13 PCGs of the related lepidopteran superfamilies. The mitogenomes of Drosophila melanogaster (U37541.1) (Lewis et al. Citation1995) and Locusta migratoria (NC_001712) (Flook et al. Citation1995) were used as outgroup. The multiple alignments of the 13 PCGs concatenated nucleotide sequences were conducted using ClustalX version 2.0 (Thompson et al. Citation1997). Then, concatenated set of nucleotide sequences from the 13 PCGs was used for phylogenetic analyses, which were performed using Maximum Likelihood (ML) method with the MEGA X program. The phylogenetic analysis reveals that Zygaenoidea, Noctuoidea, Geometroidea, Bombycoidea, Pyraloidea, Papilionoidea, Yponomeutoidea, and Hepialoidea families are monophyletic, and Zygaenoidea is more closely related to the Pyraloidea superfamiliy. The different species of same family forms a single cluster, moreover P. tessellata was found closely related to P. consocia of the Zygaenoidea (). The results are consistent with previous studies. It is concluded from the results that further studies are required to confirm the phylogentic relationships among these superfamiles.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Dai L, Qian C, Zhang C, Wang L, Wei G, Li J, Zhu B, Liu C. 2015. Characterization of the complete mitochondrial genome of Cerura menciana and Comparison with Other Lepidopteran Insects. PLoS One. 10:e0132951.
- Flook PK, Rowell CH, Gellissen G. 1995. The sequence, organization, and evolution of the Locusta migratoria mitochondrial genome. J Mol Evol. 41:928–941.
- Lewis DL, Farr CL, Kaguni LS. 1995. Drosophila melanogaster mitochondrial DNA: completion of the nucleotide sequence and evolutionary comparisons. Insect Mol Biol. 4:263–278.
- Sun Y, Tian S, Qian C, Sun Y-X, Abbas MN, Kausar S, Wang L, Wei G, Zhu B-J, Liu C-L. 2016. Characterization of the complete mitochondrial genome of Spilarctia robusta (Lepidoptera: Noctuoidea: Erebidae) and its phylogenetic implications. Eur J Entomol. 113:558–570.
- Sun YU, Zhang J, Li Q, Liang D, Abbas MN, Qian C, Wang L, Wei G, Zhu BJ, Liu CL. 2017. Mitochondrial genome of Abraxas suspecta (Lepidoptera: Geometridae) and comparative analysis with other Lepidopterans. Zootaxa. 4254:501–519.
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876–4882.
- Wu QL, Liu W, Shi BC, Gu Y, Wei SJ. 2013. The complete mitochondrial genome of the summer fruit tortrix moth Adoxophyes orana (Lepidoptera: Tortricidae). Mitochondrial DNA. 24:214–216.