Abstract
Scutellaria baicalensis is the important herb and perennial medicinal plant, which has a medical history of thousands years in China. In this study, we assembled and annotated the complete chloroplast genome of S. baicalensis. The chloroplast genome size is 152,301 bp, containing a large single-copy region (83,521 bp), a small single-copy region (17,476 bp), and a pair of IR regions (25,652 bp). The overall nucleotide composition is: A of 30.4%, T of 31.2%, C of 19.5%, and G of 18.9%, with a total GC content of the chloroplast genome 38.4% and AT of 61.6%. Scutellaria baicalensis whole chloroplast genome contains 132 genes, including 88 protein-coding genes (PCG), 36 transfer RNA (tRNAs), and eight ribosome RNA (rRNAs). Genetic and Phylogenetic analysis based on 28 herbal medicine plants species confirmed the position of S. baicalensis closely related to two other Scutellaria species of Scutellaria insignis and Scutellaria lateriflora. The complete chloroplast genome of S. baicalensis study is able to provide a reference for the phylogenetic relationships and assessment of the genetic structure of the Scutellaria family in China.
Scutellaria baicalensis is one of the most commonly used medicines in China. It is important that 70% of Chinese patent medicines and prescriptions clearing away heat dampness and purging fire detoxification has S. baicalensis medicinal ingredients (Huynh et al. Citation2017). It is widely distributed in northern China and adjacent areas of Japan, Korea, Mongolia, and Russia (Zhao et al. Citation2016). Flavones is one of the main active constituents and such as baicalin, wogonoside and others from S. baicalensis, which has anti-cancer, hepatoprotection, antibacterial antiviral and antioxidant (Woźniak et al. Citation2004). Now, we have less knowledge about the chloroplast genome of the S. baicalensis and other Scutellaria family herb plants. So, in this review, we assembled and annotated the complete chloroplast genome of S. baicalensis and discussed genetic and phylogenetic relationship with other herbal medicine plants, which contributes to study the S. baicalensis and other Scutellaria family plants genetic diversity and genetic structure of natural populations.
The specimen sample of S. baicalensis was isolated and deposited from The First People's Hospital of Jinan (Jinan, Shandong, China, 117.03E; 36.68N). The whole plant tissue and total genomic DNA was extracted using Plant Tissues Genomic DNA Extraction Kit (TaKaRa, DL, CN) and stored in The First People's Hospital of Jinan (Number TFPHJN001). The whole genomic DNA was purified and fragmented using the NEB Next Ultra™ II DNA Library Prep Kit (NEB, BJ, CN), after that the whole genomic was sequenced using the Illumina HiSeq 4000 Sequencing Platform (Illumina, San Diego, CA). Quality reads and adapters control was performed and removed low-quality reads and adapters using the NGS QC Toolkit software (Patel and Jain Citation2012). The chloroplast genome was assembled and annotated using the MitoZ software (Meng et al. Citation2019). The physical map of the new chloroplast genome was generated using OGDRAW (Lohse et al. Citation2013).
The whole complete chloroplast genome of S. baicalensis (GenBank accession No. MH6863231) was a circle with 152,301 bp in size, containing a large single-copy region (LSC) of 83,521 bp, a small single-copy region (SSC) of 17,476 bp, and a pair of inverted repeat regions (IRA and IRB) of 25,652 bp. The cpDNA of S. baicalensis comprised 132 genes, including 88 protein-coding genes (PCG), 36 transfer RNA genes (tRNA), and eight ribosomal RNA genes (rRNA). In the IR regions, a total of 18 genes were found duplicated, including seven PCG species (rpl2, rpl23, ycf2, ycf15, ndhB, rps7, and rps12), seven tRNA species (trnI-CAU, trnL-CAA, trnV-GAC, trnI-GAU, trnA-UGC, trnR-ACG, and trnN-GUU) and 4 rRNA species (rrn16, rrn23, rrn4.5, and rrn5). The overall nucleotide composition is 30.4% A, 31.2% T, 19.5% C, and 18.9% G, with a total GC content of 38.4% and AT of 61.6%.
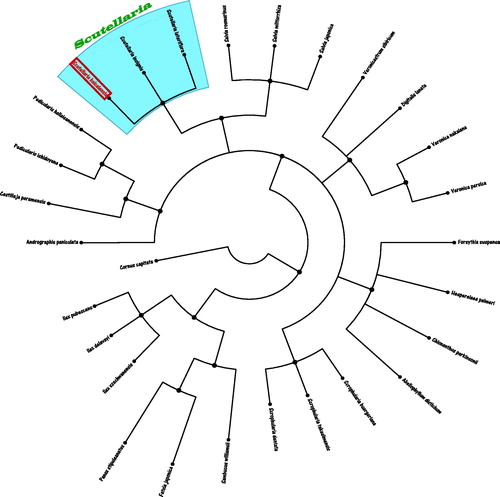
Genetic and phylogenetic analysis using the Maximum-Likelihood (ML) methods, we selected and analyzed the relationship of other 27 herbal medicine plant species chloroplast genomes with S. baicalensis. The genetic and phylogenetic tree was reconstructed using Maximum-Likelihood (ML) methods. ML analysis was performed using RAxML software (Stamatakis Citation2014) with the GTR + G + I model. Branch support was inferred using 2000 bootstrap replicates. Phylogenetic relationship obtained with the ML approach were identical to those obtained using the Bayesian analysis. The Bayesina phylogenetic alignment was analyzed using MrBayes version 3.2.5 software (Ronquist and Huelsenbeck Citation2003) based on the most appropriate model. The phylogenetic tree was represented using MEGA X software (Kumar et al. Citation2018) and edited using FigTree version 1.4.4 software. As shown in the phylogenetic ML tree result (), the chloroplast genome of S. baicalensis is clustered and closest to two other Scutellaria species of S. insignis (KT750009.1) and S. lateriflora (KY085900.1) in the genetic evolutionary relationship.
Figure 1. The Neighbour-Joining (NJ) phylogenetic tree from 28 species chloroplast genomes of herbal medicine plants. The length of branch represents the divergence distance. All the herbal medicine species chloroplast genomes in this study have been deposited in the GenBank and accession numbers are as follows: Abeliophyllum distichum (KT274029.1), Andrographis paniculata (KF150644.1), Castilleja paramensis(KT959111.1), Chionanthus parkinsonii (MG255752.1), Cornus capitate (MG524990.1), Digitalis lanata (KY085895.1), Fatsia japonica (KR021045.1), Forsythia suspense (MF579702.1), Hesperelaea palmeri (LN515489.1), Ilex delavayi (KX426470.1), Ilex pubescens (KX426467.1), Ilex szechwanensis (KX426466.1), Panax stipuleanatus (KX247147.1), Pedicularis hallaisanensis (MG770330.1), Pedicularis ishidoyana (KU170194.1), Salvia japonica (KY646163.1), Salvia miltiorrhiza (HF586694.1), Salvia rosmarinus (KR232566.1), Sambucus williamsii (KX510276.1), Scrophularia buergeriana (KP718626.1), Scrophularia dentate (MF861202.1), Scrophularia takesimensis (KM590983.1), Scutellaria insignis (KT750009.1), Scutellaria lateriflora (KY085900.1), Veronica nakaiana (KT633216.1), Veronica persica (KT724052.1), and Veronicastrum sibiricum (KT724053.1).
Acknowledgments
No project pillar.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Huynh DL, Sharma N, Kumar Singh A, Singh Sodhi S, Zhang JJ, Mongre RK, Ghosh M, Kim N, Ho Park Y, Kee Jeong D. 2017. Anti-tumor activity of wogonin, an extract from Scutellaria baicalensis, through regulating different signaling pathways. Chinese J Nat Med. 15:15–40.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35:1547–1549.
- Li H, Jiang Y, Chen F. 2004. Separation methods used for Scutellaria baicalensis active components. J Chromatogr B Analyt Technol Biomed Life Sci. 812:277–290.
- Lohse M, Drechsel O, Kahlau S, Bock R. 2013. OrganellarGenomeDRAW-a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 41:W575–W581.
- Meng G, Yiyuan L, Chentao Y, Shanlin L. 2019. MitoZ: a toolkit for animal mitochondrial genome assembly, annotation and visualization. Nucleic Acids Res. doi: 10.1093/nar/gkz173.
- Patel RK, Jain M. 2012. NGS QC Toolkit: a toolkit for quality control of next generation sequencing data. PLoS One. 7:e30619.
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 19:1572–1574.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Woźniak D, Lamer-Zarawska E, Matkowski A. 2004. Antimutagenic and antiradical properties of flavones from the roots of Scutellaria baicalensis Georgi. Mol Nutr Food Res. 48:9–12.
- Zhao Q, Chen XY, Martin C. 2016. Scutellaria baicalensis, the golden herb from the garden of Chinese medicinal plants. Sci Bull. 61:1391–1398.
