Abstract
Nilaparvata lugens is one of important pests of rice causing severe damagein early September in Korea after migrating from China. We completed mitogenome of N. lugens captured in Korea. The circular mitogenome of N. lugens is 17,610 bp including 13 protein-coding genes, 2 rRNAs, 22 tRNAs, and a 2424-bp--non-coding region. The base composition was AT-biased (89.5%). 112 single nucleotide polymorphisms (SNPs) and 59 insertions and deletions are identified by comparing with Chinese N. lugens. Phylogenetic trees presented an incongruence of topology in N. lugens clade, suggesting a need for further analyses to classify biotypes based on complete mitochondrial genomes.
Nilaparvata lugens, called brown planthoppers, is one of the important pests affecting rice (Sogawa and Cheng Citation1979). During high infestations, brown planthopper populations cause ‘hopper burn,’ lower infestations reduce the number of tillers and panicles of rice, resulting in the loss of total weight of grains (Bae and Pathak Citation1970). Nilaparvata lugens is widely distributed in rice cultivated regions including Asia, Northern Australia, and South Pacific archipelago (Hinckley Citation1963; Abraham and Nair Citation1975). Three biotypes (biotype 1, 2, and 3), of which mitochondrial genomes were already sequenced (Lv et al. Citation2015), were distinguished by preference of rice cultivars, TN1, Mudgo, and ASD7, respectively (Claridge and Den Hollander Citation1980; Diehl and Bush Citation1984; Tang et al. Citation2010). Brown planthoppers migrating from China to Korea can infect rice fields in early September (Kim et al. Citation1985; Saxena and Barrion Citation1985). To control brown planthoppers in fields effectively, its genetic background is important.
With already published mitochondrial genomes of N. lugens (Zhang et al. Citation2013; Lv et al. Citation2015), whole mitochondrial genome of N. lugens captured at Hadong-gun, Gyoungsang-nam province, the Republic of Korea (34°58′34.55ʺN, 127°50′56.40ʺE; Specimen was deposited at InfoBoss Herbarium (IN), Korea; Choi NJ, INH-00017), was completed. Total DNA was extracted using a manual DNA extraction method (Zymo Research, USA). Sequencing was conducted by HiSeqX (Macrogen Inc., Korea) and de novo assembled and confirmation were done by Velvet 1.2.10 (Zerbino and Birney Citation2008), SOAPGapCloser 1.12 (Zhao et al. Citation2011), BWA 0.7.17 (Li Citation2013), and SAMtools 1.9 (Li et al. Citation2009). Geneious R11 11.1.5 (Biomatters Ltd, Auckland, New Zealand) was used to annotate mitochondrial genome based on N. lugens mitochondrial genome (NC_021748; referred as reference genome; Zhang et al. Citation2013) together with ARWEN for identifying tRNAs (Laslett and Canbäck Citation2008).
Nilaparvata lugens mitochondrial genome (Genbank accession is MK590088) is 17,610 bp long, which is shorter than the reference genome by 10 bp. It contains 13 protein-coding genes, 2 rRNAs, and 22 tRNAs. Its nucleotide composition is AT-biased (A + T ratio is 77.1%). The control region, presumably the single largest non-coding AT-rich region, is 2,424 bp (AT ratio is 80.7%). 112 single nucleotide polymorphisms (SNPs) and 59 insertions and deletions are identified against the reference genome, which is lower than that of Chilo suppressallis (Park et al. Citation2019). Fourteen synonymous SNPs in nine genes and one non-synonymous SNP in ND5 are found.
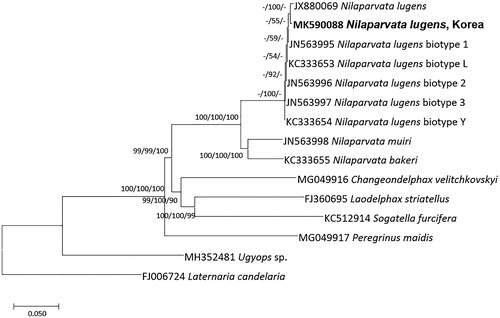
We inferred phylogenetic relationship from 14 Delphacidae and 1 Fulgoridae complete mitochondrial genomes. They were aligned by MAFFT 7.388 (Katoh and Standley Citation2013). Neighbor joining (bootstrap repeat is 10,000), minimum evolution (bootstrap repeat is 10,000), and maximum likelihood (bootstrap repeat is 1,000) trees were constructed using MEGA X (Kumar et al. Citation2018; ). Phylogenetic trees present that Korean N. lugens is clustered with brown planthopper captured in Hainan, China rather than five biotypes of N. lugens isolated in China (); but topology of N. lugens clade is not congruent among three phylogenetic trees (). It reflects that Korean N. lugens may have different mitochondrial genotypes from five biotypes, suggesting a need for further analyses to classify biotypes based on mitochondrial genomes.
Figure 1. Neighbor joining (bootstrap repeat: 10,000), minimum evolution (bootstrap repeat: 10,000), and maximum likelihood (bootstrap repeat: 1,000) phylogenetic trees of 14 Delphacidae and 1 Fulgoridae complete mitochondrial genomes, with seven species: Nilaparvata lugens (MK590088 in this study, JX880069, JN563995, KC333653, JN563996, JN563997, and KC333654), Nilaparvata muiri (JN563998), Nilaparvata bakeri (KC333655), Changeondelphax velitchkovskyi (MG049916), Laodelphax striatellus (FJ360695), Sogatella furcifera (KC512914), Peregrinus maidis (MG049917), Ugyops sp. (MH352481), and Laternaria Candelaria (FJ006724), as outgroup species. Phylogenetic tree was shown based on minimum evolution tree. The numbers above branches indicate bootstrap support values of neighbor joining, minimum evolution, and maximum likelihood phylogenetic trees, respectively.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Abraham C, Nair M. 1975. The brown planthopper outbreaks in Kerala, India: Rice Entomol Newsl. 2:36.
- Bae SH, Pathak M. 1970. Life history of Nilaparvata lugens (Homoptera: Delphacidae) and susceptibility of rice varieties to its attacks. Ann Entomol Soc Am. 63:149–155.
- Claridge MF, Den Hollander J. 1980. The “biotypes” of the rice brown planthopper, Nilaparvata lugens. Entomol Exp Applic. 27:23–30.
- Diehl S, Bush GL. 1984. An evolutionary and applied perspective of insect biotypes. Ann Rev Entomol. 29:471–504.
- Hinckley AD. 1963. Ecology and control of rice planthoppers in Fiji. Bull Entomol Res. 54:467–481.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Kim Y, Lee J, Park H, Kim M. 1985. Plant damages and yields of the different rice cultivars to brown planthopper (Nilaparvata lugens S.) in fields. Kor J Appl Entomol. 24:79–83.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol Biol Evol. 35:1547–1549.
- Laslett D, Canbäck B. 2008. ARWEN: a program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics. 24:172–175.
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. Genomics. 1303.
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The sequence alignment/map format and SAMtools. Bioinformatics. 25:2078–2079.
- Lv L, Peng X, Jing S, Liu B, Zhu L, He G. 2015. Intraspecific and interspecific variations in the mitochondrial genomes of Nilaparvata (Hemiptera: Delphacidae). J Econ Entomol. 108:2021–2029.
- Park J, Xi H, Kwon W, Park C-G, Lee W. 2019. The complete mitochondrial genome sequence of Korean Chilo suppressalis (Walker, 1863) (Lepidoptera: Crambidae). Mitochondrial DNA Part B. 4:850–851.
- Saxena R, Barrion A. 1985. Biotypes of the brown planthopper Nilaparvata lugens (Stål) and strategies in deployment of host plant resistance. Int J Trop Insect Sci. 6:271–289.
- Sogawa K, Cheng C. 1979. Brown planthopper: threat to rice production in Asia. Manila: International Rice Res Institute. Economic thresholds, nature of damage, and losses caused by the brown planthopper; p. 125–142.
- Tang M, Lv L, Jing S, Zhu L, He G. 2010. Bacterial symbionts of the brown planthopper, Nilaparvata lugens (Homoptera: Delphacidae). Appl Environ Microbiol. 76:1740–1745.
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Gen Res. 18:821–829.
- Zhang K-J, Zhu W-C, Rong X, Zhang Y-K, Ding X-L, Liu J, Chen D-S, Du Y, Hong X-Y. 2013. The complete mitochondrial genomes of two rice planthoppers, Nilaparvata lugens and Laodelphax striatellus: conserved genome rearrangement in Delphacidae and discovery of new characteristics of atp8 and tRNA genes. BMC Genomics. 14:417.
- Zhao Q-Y, Wang Y, Kong Y-M, Luo D, Li X, Hao P. 2011. Optimizing de novo transcriptome assembly from short-read RNA-Seq data: a comparative study. BMC bioinformatics. 12:S2.
