Abstract
According to the reference data in GenBank, the complete mitochondrial genome KP281293.1 presumably belongs to Hypomesus olidus from China. The phylogenetic analysis based on the Cytb and CoI genes of the smelt genus Hypomesus suggests that the Chinese specimen belongs to Hypomesus nipponensis. The difference in the studied region of mitogenome is low, 0.6%, which is in agreement with the values of intraspecific divergence for the smelts. The use of the sequence KP281293.1 as a marker for H. olidus will not help in improving the understanding of the taxonomic relationships gained from previous morphological studies and is phylogenetically misleading.
The geographic distribution pattern of mtDNA sequence polymorphisms of H. olidus Pallas 1814, over most of the range in the Northwestern Pacific, was previously examined (Skurikhina et al. Citation2012). Having studied more samples, we found a mistake in the taxonomic identification of the complete mitochondrial genome of H. olidus from Bai et al. (Citation2017). Incomplete phylogeny of the family Osmeridae has been recently published, including the mitogenomes of this smelt (Balakirev et al. Citation2018). However, taxonomic errors can lead to inaccurate phylogenetic conclusions. In the present study, we would like to warn the investigators against such mistakes.
The specimen KP281293.1 has a taxonomy ID (NCBI: txid240830) and presumably belongs to H. olidus from Yuqiao Reservoir, Tianjin, China (40°01′ N/117°54′ E). To explore the phylogenetic position of this specimen, phylogenies involving other smelt species were constructed using public Cytb and CoI genes sequences. 119 specimens of H. olidus from nine different locations were included to place our findings within a broader phylogenetic group. These include all specimens of H. olidus reported by Skurikhina et al. (Citation2012) and Melnikova et al. (Citation2018). All sequences were obtained from the Genbank/NCBI database (HQ115079-HQ115271, FJ205570-FJ205572, FJ010869-FJ010871, MK038781-MK038784, MK038788-MK038807, MK038813-MK038816, MK038818-MK038837, and KY400645-KY400647). The samples of smelts H. nipponensis and H. japonicus from the Olga Bay of the Sea of Japan (43°43′ N/135°15′ E) were included for comparison. The fish specimens are stored in the collection of Genetics Laboratory, National Scientific Center of Marine Biology, Vladivostok, Russia (www.imb.dvo.ru). The complete mitochondrial genome of H. nipponensis (HM106489.1) from Jingpo Lake, China (43°52′ N/128°56′ E) was added to the analysis. The total length of mtDNA nucleotide sequences was 1668 bp.
As shown in the maximum-likelihood tree (), the specimen KP281293.1 was clustered with H. nipponensis with a very low level of sequence divergence (Dxy = 0.006 ± 0.002), while all sequences of H. olidus proper formed a different cluster. The low level of sequence divergence was also detected within the group of H. olidus (0.0001 ± 0.0001 – 0.0024 ± 0.0010), significantly lower than that between KP281293.1 and H. olidus (0.1296 ± 0.0085).
Figure 1. Maximum likelihood tree for the smelt specimen KP281293.1 (NC_026566.1) and GenBank representatives of the smelt genus Hypomesus as inferred from the concatenated sequence of Cytb and CoI genes. The tree is based on the TrN93 plus gamma model of nucleotide substitution. Numbers at the nodes indicate bootstrap probabilities from 1000 replications (values below 80% are omitted). The tree was rooted with three Osmerus dentex haplotypes.
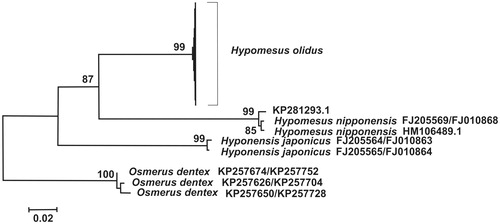
Our results suggest that the specimen KP281293.1 belongs to H. nipponensis. With respect to Hypomesus phylogeny, the relationships between H. olidus and H. nipponensis are not yet clear (Ilves and Taylor Citation2009; Skurikhina et al. Citation2013). In this situation, the inclusion of the complete mitogenome KP281293.1 may cause a conflicting phylogenetic signal. The arguments for the erroneous taxonomic assignment by Bai et al. (Citation2017) are the following. Of the three species (H. japonicus, H. nipponensis, and H. olidus) inhabiting the Sea of Japan basin, H. olidus is only found in its northern part (Chereshnev et al. Citation2001). We suppose that the specimen KP281293.1 was caught in China, where H. olidus is never found in natural populations.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Bai X, Luo Z, Feng S, Sun Z, Wu HM, Li CY, Jiang JF, Liu X, Wang N. 2017. The complete mitochondrial genome of Hypomesus olidus (Osmeriformes: Osmeridae). Mitochondrial DNA (Part B). 2:687–688.
- Balakirev ES, Romanov NS, Ayala FJ. 2018. Complete mitochondrial genome of the surf smelt Hypomesus japonicus (Osmeriformes, Osmeridae). Mitochondrial DNA (Part B). 3:1071–1072.
- Chereshnev IA, Shestakov AV, Frolov SV. 2001. On the systematics of species of the genus Hypomesus (Osmeridae) of Peter the Great Bay, Sea of Japan. Rus J Marine Biol. 27:296–302.
- Ilves KL, Taylor EB. 2009. Molecular resolution of the systematics of a problematic group of fishes (Teleostei: Osmeridae) and evidence for morphological homoplasy. Mol Phylogenet Evol. 50:163–178.
- Melnikova MN, Senchukova AL, Pavlov SD. 2018. New data about mtDNA variability of pond smelt Hypomesus olidus (Osmeridae) from the Commander Islands in comparison with other populations of the species. Biol Bull Russ Acad Sci. 45:11–17.
- Skurikhina LA, Kukhlevsky AD, Kovpak NE. 2013. Relationships of osmerid fishes (Osmeridae) of Russia: divergence of nucleotide sequences of mitochondrial and nuclear genes. Genes Genom. 35:529–539.
- Skurikhina LA, Kukhlevsky AD, Zheleznova KO, Kovalev MY. 2012. Analysis of the mitochondrial DNA variation in pond smelt Hypomesus olidus (Osmeridae). Russ J Genet. 48:713–722.
