Abstract
We used next-generation sequencing to determine the complete chloroplast genome of Fagus multinervis (Fagaceae), a beech endemic to Ulleung Island in South Korea. This genome is 158,348 bp long, has a typical quadripartite structure, and contains a large single copy region (87,659 bp), small single copy region (18,903 bp), and two inverted repeats (25,893 bp). Overall GC content is 37.1%. The genome encodes 129 genes, including 83 for proteins, 38 for tRNA, and 8 for rRNA. Based on a comparison of 63 protein-coding genes from related species, we placed Fagus at a basal position on our phylogenetic tree.
Genus Fagus L. (Fagaceae Dumort.) comprises 10 tree species distributed within temperate zones in the Northern Hemisphere (Huang et al. Citation1999). Among those found in Korea, China, and Japan, seven species are highly valued for their timber (Huang et al. Citation1999; Chang Citation2007; Yu Citation2016). Fagus multinervis Nakai is endemic and limited to Ulleung Island in South Korea (Chang Citation2007). Its current taxonomic status is controversial. For example, Lee (Citation1967) includes it in F. japonica Maxim in Japan, based on morphological characters. In China, however, Shen (Citation1992) treats it as a synonym of F. engleriana Seemen, again based on morphological similarities. Nevertheless, the bark pattern is vertically elongated for both F. multinervis and F. japonica, a characteristic distinct from that of F. engleriana. Furthermore, the cupules from both F. multinervis and F. japonica are covered with leaflike bracts that differ from those of F. engleriana. A previous molecular phylogenetic study of F. multinervis and those two related species (Oh et al. Citation2016) demonstrated that, although F. multinervis shows monophyly, the relationship among the three species remains unclear, and data suggest that F. multinervis originated through hybridization. Here, we report the complete chloroplast (cp) genome of F. multinervis in an effort to resolve this argument within Fagus.
Total genomic DNA was extracted from dry leaves of Fagus multinervis collected on Ulleung Island (Voucher specimen: 37°30′ N, 130°54′ E, J. S. Park & D. P. Jin 185109, KH: Korea National Arboretum). This genomic DNA was sequenced with the Illumina MiSeq platform (Macrogen, Seoul, Korea). The 10,007,240 paired reads were then assembled using the cp of F. engleriana from China as our reference genome (GenBank: KX852398) (Yang et al. Citation2018). For the draft genome, we employed Sanger sequencing to validate four junctions between the large single copy (LSC) region, small single copy (SSC) region, and two inverted repeats (IRs), and regions showing low coverage (<100). Annotation was conducted with tRNAscan-SE (Lowe and Chan Citation2016), using this draft genome along with cp genomes from F. engleriana and Quercus acutissima Carruth. (GenBank: MF593895). Our analysis indicated that the cp genome of F. multinervis (GenBank: MK518070) is 158,348 bp long. It consists of an LSC region (87,659 bp), SSC region (18,903 bp), and a pair of IRs (25,893 bp). Overall GC content is 37.1% (LSC, 35.0%; SSC, 31.1%; IRs, 42.7%). This cp genome encodes 129 genes – 83 protein-coding, 38 for tRNA, and 8 for rRNA. As it is known from F. engleriana, three protein-coding genes (rps16, infA, and rpl22) have been lost from F. multinervis (Yang et al. Citation2018).
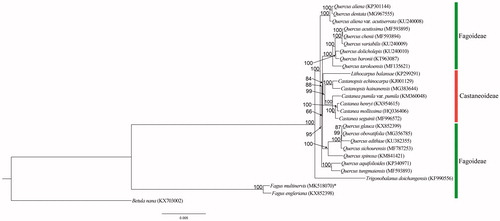
Based on the known cp genomes from 26 species, we constructed a phylogenetic tree for Fagaceae (). In all, we used 63 protein-coding genes to generate a Maximum likelihood tree with RAxML 8.2.11 (Stamatakis Citation2014). Betula nana L. (GenBank: KX703002) of Betulaceae served as the outgroup. We placed Fagus at the basal position. This tree revealed that, except for Quercus, the genera within Fagaceae form a monophyletic group, as suggested by Yang et al. (Citation2018).
Disclosure statement
The authors report no conflicts of interest, and all are responsible for the content and writing of this article.
Additional information
Funding
References
- Chang C-S. 2007. Fagus L. In: Park CW, editor. The genera of vascular plants of Korea. Seoul, South Korea: Academy Publishing Co.; p. 268–269.
- Huang C, Zhang Y, Bartholomew B. 1999. Fagus L. In: Flora of China Editorial Committee, editor. Flora China Vol. 4. Beijing and St. Louis: Science Press and Missouri Botanical Garden Press.; p. 314–315.
- Lee YN. 1967. Taxonomic studies on the Fagus multinervis, Fagus japonica and Fagus crenata. Bull Korean Inst Cult Res. 10:373–377.
- Lowe TM, Chan PP. 2016. tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44:W54–W57.
- Oh S-H, Youm J-W, Kim Y-I, Kim Y-D. 2016. Phylogeny and evolution of endemic species on Ulleungdo Island, Korea: The case of Fagus multinervis (Fagaceae). Syst Bot. 41:617–625.
- Shen C-F. 1992. A monograph of the genus Fagus Tourn. ex L. (Fagaceae) [dissertation]. New York City (NY): City University of New York.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Yang Y, Zhu J, Feng L, Zhou T, Bai G, Yang J, Zhao G. 2018. Plastid genome comparative and phylogenetic analyses of the key genera in Fagaceae: highlighting the effect of codon composition bias in phylogenetic inference. Front Plant Sci. 9:1–13.
- Yu I. 2016. Fagus L. In: Ohashi H, Kadota Y, Murata J, Yonekura K, Kihara H, editors. Wild flowers of Japan Vol. 3. Tokyo, Japan: Heibonsha; p. 91–93.