Abstract
The Cape petrel Daption capense, a seabird, is observed throughout the southern hemisphere including the Antarctic region. Herein, we report the complete mitogenome of Cape petrel with the GenBank accession number MH924023 to elucidate the genetic characteristics of this genus. The mitogenome is 17,312 bp long and comprises 13 protein-coding genes (PCGs), two rRNA genes, and 22 tRNA genes. The overall base composition of the 13 PCGs is as follows: A (31.1%), C (30.3%), T (25.4%), and G (13.2%), with a GC content of 43.5%. The results provide useful information for further genetic studies in Procellariiformes.
Cape petrel Daption capense, a seabird distributed throughout the Southern Ocean, belongs to the family Procellariidae and order Procellariiformes, is the only species in the genus Daption and has two morphologically distinct subspecies, viz., D. c. capense and D. c. australe. Although the population of Cape Petrel is decreasing in some areas (Petry et al. Citation2018), the global population is stable (BirdLife International Citation2019). There is still a controversy over classification of Procellariiformes using partial mitochondrial DNA such as cytochrome B (Penhallurick and Wink Citation2004; Rheindt and Austin Citation2005); thus, there is a need for basic genetic information on various species of this order to understand their phylogeny and evolution.
A Cape Petrel carcass was collected in the southern part of Barton Peninsula (62°14′33.31″S, 58°43′45.26″W), King George Island, Antarctica on January 3, 2011 and transferred to the King Sejong Station; muscle tissue (sample no. JHK-56) was separated from the carcass and frozen at −20 °C until further use. The specimen was stored in the refrigerator of the Korea Polar Research Institute (KOPRI) at Incheon, South Korea. Genomic DNA was extracted using the DNeasy Blood and Tissue Kit (Qiagen, Hilden, Germany). Sequencing and assembly were conducted as previously reported by Lee et al. Citation(2018). High-quality reads of 1.59 Gbp were obtained from paired-end raw reads and de novo assembled using CLC Assembly Cell Package (ver. 4.2.1, CLC Bio Inc., Aarhus, Denmark). Mitochondrial contigs were selected from the assembled contigs and merged into a draft genome sequence, which was manually confirmed and edited by paired-end read mapping. The mitochondrial genes were primarily annotated using GeSeq (Tillich et al. Citation2017). Accurate gene annotation was determined using the Artemis annotation tool (Rutherford et al. Citation2000) and BLASTN searches against the National Center for Biotechnology Information organelle genome database. The 12 mitochondrial PCGs (atp6, atp8, cytb, cox1, cox2, cox3, nd2, nd3, nd4, nd4l, nd5, and nd6) in 13 species from the class Aves were concatenated and aligned using MAFFT (version 7; Katoh and Standley Citation2013). Bayesian inference analysis was performed with GTR + Γ + I model using MrBayes v3.2.6 (Ronquist et al., Citation2012). Four Markov chain Monte Carlo were run for 10,000,000 generations and a burn-in of 0.25.
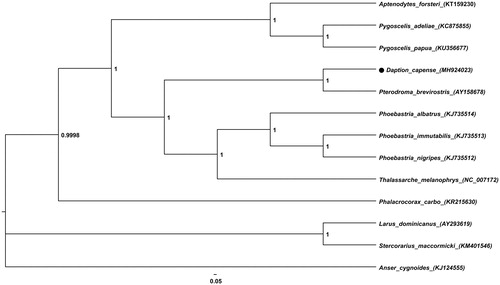
We found that the D. Capense mitogenome comprises 17,312 bp (GenBank accession number MH924023) and includes 13 protein-coding genes (PCGs), 2 rRNA genes, and 22 tRNA genes. The overall base composition is as follows: 31.1% (A), 25.4% (T), 30.3% (C), and 13.2% (G) with a 43.5% GC content. Further, a heterogeneous region of a single base (1019 bp), composed of 60.33% T and 39.67% C bases (depth ratio 73/48), was found in silico and we considered it as a T-base region. The phylogenetic tree of D. capense with 12 Aves species was constructed using FigTree v1.4.3 (Rambaut, Citation2016) ( ). Currently, only five sequences of Procellariiformes are available in the GenBank and, therefore, genetic studies on Procellariiformes are limited. The mitogenome of D. capense is expected to provide basic data for more accurate classification of this order.
Figure 1. Bayesian inference phylogeny of Daption capense with 12 Aves species based on the commonly conserved 12 mitochondrial protein-coding sequences. Posterior probability was calculated using MrBayes program. GenBank accession numbers are presented next to the species names. Anser cygnoides was used as the outgroup.
Disclosure statement
We declare no potential conflict of interest.
Additional information
Funding
References
- BirdLife International. 2019. Species factsheet: Daption capense. [accessed 2019 Feb 19]. http://www.birdlife.org.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Lee, H-O, Choi, J-W, Baek, J-H, Oh, J-H, Lee, S-C, Kim, C-K. 2018. Assembly of the Mitochondrial Genome in the Campanulaceae Family Using Illumina Low-Coverage Sequencing. Genes. 9:383 DOI:10.3390/genes9080383
- Penhallurick J, Wink M. 2004. Analysis of the taxonomy and nomenclature of the Procellariiformes based on complete nucleotide sequences of the mitochondrial cytochrome b gene. Emu. 104:125–147.
- Petry MV, Valls FCL, Petersen ES, Finger JVG, Krüger L. 2018. Population trends of seabirds at Stinker Point, Elephant Island, Maritime Antarctica. Antarctic Sci. 30:220–226.
- Rambaut, A. 2016. Figtree v1.4.3. [accessed2019 March 26]. Available from: http://tree.bio.ed.ac.uk/software/figtree/.
- Rheindt FE, Austin JJ. 2005. Major analytical and conceptual shortcomings in a recent taxonomic revision of the Procellariiformes – a reply to Penhallurick and Wink (2004). Emu. 105:181–186.
- Ronquist, F, Teslenko, M, van der Mark, P, Ayres, D L, Darling, A, Höhna, S, Larget, B, Liu, L, Suchard, M A, Huelsenbeck, J P. 2012. MrBayes 3.2: Efficient Bayesian Phylogenetic Inference and Model Choice Across a Large Model Space. Systematic Biology. 61:539–542. DOI:10.1093/sysbio/sys029
- Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream MA, Barrell B. 2000. Artemis: sequence visualization and annotation. Bioinformatics. 16:944–945.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45:W6–W11.
