Abstract
Gansu wapiti (Cervus elaphus kansuensis) is one of eight subspecies of wapiti in China, which has been placed under the second-ranked protected animals by Chinese government. No complete mitochondrial genomes of Gansu wapiti was determined until now, so the phylogenetic relationships among the subspecies of wapiti and other species of the genus Cervus have not been well studied. In this study, the complete mitochondrial genome of C. e. kansuensis was first sequenced and characterized. The genome is 16,430 bp in length, containing 13 protein-coding genes (PCGs), 22 transfer RNA (tRNA) genes, two ribosomal RNA (rRNA) genes, and one noncoding control region (CR). Maximum-Likelihood (ML) and Bayesian Inference (BI) trees based on the 13 PCGs show that C. e. kansuensis is clustered with other wapiti subspecies in China except for C. e. yarkandensis. It is also shown that Cervus elaphus is not a monophyletic group. This study provides important molecular evidence for the phylogenetic relationship among species in the genus Cervus.
Wapiti is naturally distributed throughout Asia and North America and listed as a least concern species by the International Union for Conservation of Nature and Natural Resources (Brook et al. Citation2018), which have been placed under the second-ranked protected animals by Chinese government since 1989. Gansu wapiti (Cervus elaphus kansuensis) is classified as one of eight wapiti subspecies in China, which occurs in Gansu, Qinghai, and northern Sichuan (Ohtaishi and Gao Citation1990). Until now, the classification information for Gansu wapiti mainly comes from traditional taxonomy according to variety of morphological differences and geographical distribution (Sheng Citation1992). The phylogenetic position of C. e. kansuensis in Cervus has not been well studied without enough molecular data.
In this study, we determined the complete mitochondrial genome of C. e. kansuensis. Blood samples were collected from a deer farm in Gansu province (38.915°N, 99.571°E). Total genomic DNA (accession No: DG1) was extracted and stored at −80 °C in Institute of Special Animal and Plant Sciences.
Nineteen pairs of primers were designed to amplify contiguous overlapping fragments. The complete mitochondrial genome of C. e. kansuensis is 16,430 bp in length (GenBank accession No: NC039923) with 62.19% A + T content. Similar to other red deer mitogenomes, it consists of 13 protein-coding genes (PCGs), 22 tRNA genes, two ribosomal RNAs and a D-loop. All PCGs are encoded on heavy strand (H-strand) except for ND6. Among 13 PCGs, 10 genes are initiated by ATG codon, whereas ATA is the initiation codon in ND2, ND3, and ND5. Eight genes are terminated with the complete stop codon (TAA for COX1, COX2, ATP8, ATP6, ND4L, ND5, and ND6 genes, AGA for CYTB genes), whereas five genes use incomplete stop codons (TA– for ND1, T–– for ND2, COX3, ND3, and ND4). Twenty-two tRNA genes were ranging from 60 bp (tRNASer) to 75 bp (tRNALeu) in size. Fourteen tRNA genes are encoded on H-strand and eight tRNA genes are encoded on light-strand (L-strand). 12S rRNA and 16S rRNA are located between two tRNA genes (tRNAPhe and tRNALeu) and separated by tRNAVal. The D-loop (991 bp) is located between tRNAPro and tRNAPhe.
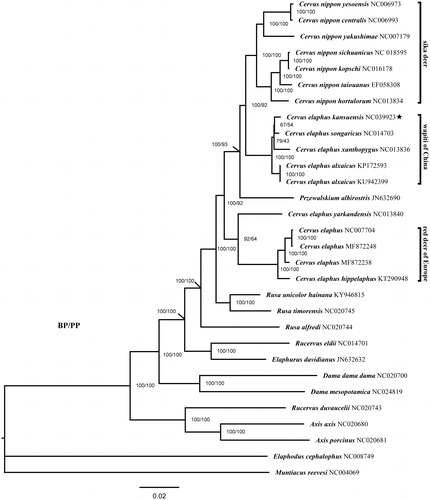
In order to illustrate the phylogenetic position of Gansu wapiti, we built phylogenetic trees based on the 13 PCGs of total 30 Cervidae species. RAxML v8 (Stamatakis Citation2014) was used for Maximum-likelihood (ML) analysis. Bayesian inference (BI) was conducted using MrBayes v3.2 (Ronquist and Huelsenbeck Citation2003). The phylogenetic analyses demonstrated that C. elaphus divide into two clades (), as many researchers found earlier (Kuwayama and Ozawa Citation2000; Randi et al. Citation2001; Ludt et al. Citation2004; Pitra et al. Citation2004; Frank et al. Citation2016). Cervus elaphus kansuensis is clustered with other wapiti subspecies in China except for C. e. yarkandensis. This clade is more closely related to the sika deer clade than to European red deer clade. The explanation for the interrelationships between sika deer and wapiti is that hybridization has taken place before wapiti spread to East Asia and North America (Groves and Grubb Citation2011). We suggest that this subspecies should be considered as an evolutionarily significant unit in the conservation of wapiti.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Brook SM, Pluháček J, Lorenzini R, Lovari S, Masseti M, Pereladova O, Mattioli S. 2018. Cervus canadensis (errata version published in 2019). The IUCN Red List of Threatened Species. 2018:e.T55997823A142396828. Downloaded on 22 March 2019.
- Frank K, Barta E, Bana NÁ, Nagy J, Horn P, Orosz L, Stéger V. 2016. Complete mitochondrial genome sequence of a Hungarian red deer (Cervus elaphus hippelaphus) from high-throughput sequencing data and its phylogenetic position within the family Cervidae. Acta Biologica Hungarica. 67:133–147.
- Groves CP, Grubb P. 2011. Ungulate Taxonomy. Baltimore: The Johns Hopkins University Press.
- Kuwayama R, Ozawa T. 2000. Phylogenetic relationships among European red deer, wapiti, and sika deer inferred from mitochondrial DNA sequences. Mol Phylogenet Evol. 15:115–123.
- Ludt CJ, Schroeder W, Rottmann O, Kuehn R. 2004. Mitochondrial DNA phylogeography of red deer (Cervus elaphus). Mol Phylogenet Evol. 31:1064–1083.
- Ohtaishi N, Gao Y. 1990. A review of the distribution of all species of deer (Tragulidae, Moschidae and Cervidae) in China. Mammal Rev. 20:125–144.
- Pitra C, Fickel J, Meijaard E, Groves PC. 2004. Evolution and phylogeny of old world deer. Mol Phylogenet Evol. 33:880–895.
- Randi E, Mucci N, Claro-Hergueta F, Bonnet A, Douzery E. 2001. A mitochondrial DNA control region phylogeny of the Cervinae: speciation in Cervus and implications for conservation. Anim Conserv. 4:1–11.
- Ronquist F, Huelsenbeck JP. 2003. MrBayes V3.2: Bayesian phylogenetic inference under mixed models. Bioinformatics. 19:1572–1574.
- Sheng H. 1992. The deer in China. Shanghai: East China Normal University Press.
- Stamatakis A. 2014. RAxML V8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.