Abstract
We sequenced the complete mitochondrial genome of Niponiella limbatella Klapálek, 1907, the first representative of the genus Niponiella, with a length of 15,924 bp. The complete mitochondrial genome of N. limbatella consists of 13 protein-coding genes (PCGs), two ribosomal RNA genes, 22 transfer RNA (tRNAs) genes, and a control region (GenBank accession No. MK686067). The nucleotide composition of N. limbatella mitochondrial genome was 33.6% of A, 29.6% of T, 23.9% of C, 13.0% of G, and 63.1% of A + T content. Initiation codon ATN was shared with eleven PCGs, except ND5 and ND1, started with codon GTG and TTG. Eleven PCGs use the conservative stop codon TAA/TAG, except COII and COIII terminated with a single T––. The phylogenetic relationships based on Bayesian (BI) and maximum-likelihood (ML) methods showed that N. limbatella is closely related to Caroperla siveci, which is in accordance with the traditional morphological classification.
Klapálek (Citation1907) established the genus Niponiella, and now only a single species has been recorded of the genus Niponiella in the world which are restricted to Japan (DeWalt et al. Citation2019). As we know, six mitochondrial genomes of Perlidae have been sequenced and published but the genus Niponiella was not involved (Qian et al. Citation2014; Elbrecht et al. Citation2015; Huang et al. Citation2015; Wang, Ding, Yang Citation2016; Wang, Wang, Yang Citation2016; Cao et al. Citation2019). Here, we sequenced and annotated the mitochondrial genome of N. limbatella Klapálek, Citation1907, the first representative of Niponiella for further study. The tissue sample of N. limbatella was collected from Gunma, Nakanojyo-machi, Japan by Liu Xingyue in 2015 (coordinate as follows: N36°704', E138°801'). Specimens are deposited in the insect specimen room of Henan Institute of Science and Technology (HIST), China with an accession number HIST-0027. The genetic DNA from adult’s chest of N. limbatella was individually extracted using the QIAamp DNA Mini Kit (QIAGEN, Hilden, Germany) following the manufacturer’s protocol and sequenced under the next generation sequence technology.
The complete mitogenome of N. limbatella is a closed-circular molecule of 15,924 bp in length and deposited in the GenBank under the accession number MK686067 in NCBI. The mitochondrial genome sequenced contains 13 protein-coding genes (PCGs), two ribosomal RNA genes (rrnL and rrnS), 22 transfer RNA genes (tRNAs), and a control region (1052 bp). The total nucleotide composition was biased towards A/T nucleotides with 33.6% of A, 29.6% of T, 23.9% of C, 13.0% of G, and 63.1% of A + T content. The mitochondrial genome showed a positive AT-skew (0.063) and a negative GC-skew (–0.295). The mitogenome contains the total of 44 bp overlapping sequences, and the longest overlap is 8 bp which was located between tRNATrp and tRNACys. This mitogenome has a total of 34 bp intergenic spacer sequences, and the largest intergenic spacer sequence (16bp) is located between tRNASer(UCN) and ND1. The length of 22 tRNAs ranged from 65 bp (tRNAGly, tRNAHis, and tRNAPro) to 72bp (tRNAVal). With an exception for tRNASer(AGN), all tRNAs have the typical cloverleaf secondary structure, which are similar to those published plecopterlid mitogenomes (Wang et al. Citation2017, Citation2019). Initiation codon of eleven PCGs of N. limbatella was shared with typical codons ATN, except ND5 and ND1 started with codon GTG and TTG. Eleven PCGs use the conservative termination codon TAA/TAG, except COII and COIII stopped with a single T––.
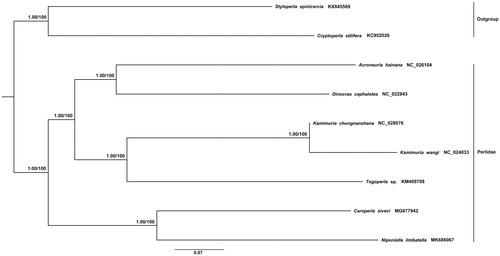
All 13 protein-coding genes (PCGs) and two Ribosomal RNAs from seven Perlidae species and two species (Cryptoperla stilifera and Styloperla spinicercia as outgroups) were used to conduct a phylogenetic analysis with the Bayesian (BI) and maximum-likelihood method (ML). Two methods showed the same topology tree which was given as . The phylogenetic tree shows that the relationship was ((Niponiella + Caroperla) + ((Acroneuria + Dinocras) + (Togoperla + Kamimuria))), and N. limbatella is closely related to Caroperla siveci, which is in accordance with the traditional morphological classification (Sivec et al. Citation1988).
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Cao JJ, Wang Y, Li N, Li WH, Chen X. 2019. Characterization of the nearly complete mitochondrial genome of a Chinese endemic stonefly species, Caroperla siveci (Plecoptera: Perlidae). Mitochondrial DNA B. 4:553–554.
- DeWalt RE, Maehr MD, Neu-Becker U, Stueber G. 2019. Plecoptera species file online. http://Plecoptera.SpeciesFile.org; [accessed 2019 Mar 25].
- Elbrecht V, Poettker L, John U, Leese F. 2015. The complete mitochondrial genome of the stonefly Dinocras cephalotes (Plecoptera, Perlidae)). Mitochondrial DNA. 26:469–470.
- Huang MC, Wang YY, Liu XY, Li WH, Kang ZH, Wang K, Li XK, Yang D. 2015. The complete mitochondrial genome and its remarkable secondary structure for a stonefly Acroneuria hainana Wu (Insecta: Plecoptera: Perlidae). Gene. 557:52–60.
- Klapálek F. 1907. Japonské druhy podčeledi Perlinae. Rozpravy České Akad. (Praha). 1631:1–28.
- Qian YH, Wu HY, Ji XY, Yu WW, Du YZ. 2014. Mitochondrial genome of the stonefly Kamimuria wangi (Plecoptera: Perlidae) and phylogenetic position of Plecoptera based on mitogenomes. PLoS One. 9:e86328.
- Sivec I, Stark BP, Uchida S. 1988. Synopsis of the world genera of Perlinae (Plecoptera: Perlidae). Scopolia. 16:1–66.
- Wang K, Ding SM, Yang D. 2016. The complete mitochondrial genome of a stonefly species, Kamimuria chungnanshana Wu, 1948 (Plecoptera: Perlidae). Mitochondrial DNA A. 27:3810–3811.
- Wang K, Wang YY, Yang D. 2016. The complete mitochondrial genome of a stonefly species, Togoperla sp. (Plecoptera: Perlidae). Mitochondrial DNA A. 27:1703–1704.
- Wang Y, Cao JJ, Li WH. 2017. The complete mitochondrial genome of the styloperlid stonefly species Styloperla spinicercia Wu (Insecta: Plecoptera) with family-level phylogenetic analyses of the Pteronarcyoidea. Zootaxa. 4243:125–138.
- Wang Y, Cao JJ, Li N, Ma GY, Li WH. 2019. The first mitochondrial genome from Scopuridae (Insecta: Plecoptera) reveals structural features and phylogenetic implications. Int J Biol Macromol. 122:893–902.