Abstract
Amentotaxus yunnanensis H. L. Li (Taxaceae) is a threatened plant species and a typical Plant Species with Extremely Small Population (PSESP). In this study, we sequenced the whole chloroplast (cp) genome of A. yunnanensis. The results showed that the plastid genome is 137,594 bp in size. In total, 118 unique genes were annotated, including 83 protein-coding genes, 31 tRNA genes, and four rRNA genes. The phylogenetic tree based on 24 cp genomes of Taxaceae support the close relationships of the genera Amentotaxus and Torreya.
Conifers have dominated forests for more than 200 million years and are very important for the ecosystem and human society (Nystedt et al. Citation2013). Taxaceae is a small family of conifers comprises of six genera (including Amentotaxus, Austrotaxus, Cephalotaxus, Pseudotaxus, Taxus, and Torreya) and about 35 species (Elpe et al. Citation2018). The genus Amentotaxus Pilg. consists of six species, one of them Amentotaxus yunnanensis H. L. Li (vernacular name Yunnan Catkin Yes) occurs in China, Laos and Vietnam and was categorized as Vulnerable (VU) in the Red List (Thomas et al. Citation2017). Moreover, this species has also been listed as a Plant Species with Extremely Small Population (PSESE) needing urgent rescue action by the local Government of Yunnan (Sun Citation2013). As a typical PSESP, A. yunnanensis is of huge ecological, scientific, cultural, and economic importance. However, this species has received very limited research and conservation attention so far. In the present study, we report and characterized the complete plastome of A. yunnanensis. The obtained results will be fundamental for the further conservation of this threatened species.
Plant material of A. yunnanensis (voucher: CL2018035) was collected from Xichou, Yunnan Province of China (23°24'19″ N, 104°50'48″ E). The individual was cultivated in Kunming Botanical Garden, Kunming Institute of Botany, CAS. An herbarium specimen was stored in Yunnan Key Laboratory for Integrative Conservation of Plant Species with Extremely Small Populations, Kunming Institute of Botany, CAS. The complete cp genome sequencing was performed on the Illumina Hiseq X platform (Illumina Inc, San Diego, CA, USA), assembled into the complete cp genome by NOVOPlasty version 2.6.2 (Dierckxsens et al. Citation2016) with A. argotaenia (Accession no.: NC_027581) as the reference. Annotated was performed using the tools DOGMA (Wyman et al. Citation2004) and plann 1.1 (Huang and Cronk Citation2015). Amentotaxus yunnanensis cp genome was deposited in Genbank under the Accession no.: MK675813.
The complete cp genome of A. yunnanensis is 137,594 bp in length. Similar to other conifers, the studied species does not contain IRs. The total GC content was 35.9%. In total, 118 unique genes were annotated, including 83 protein-coding genes, 31 tRNA genes, and four rRNA genes. Sixteen genes contained a single intron (comprising 10 protein-coding and six tRNA genes) and ycf3 encoded two introns.
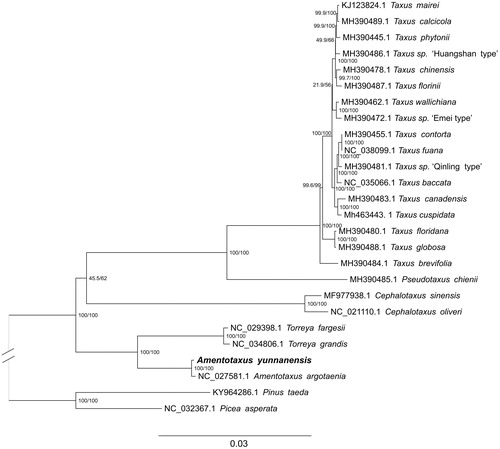
To study the phylogenetic position of A. yunnanensis 23 cp genomes from Taxaceae were selected for analyses using Pinus taeda and Picea asperata (Pinaceae clade) as outgroup. The genome-wide alignment of all cp genomes was done by HomBlocks (Bi et al. Citation2018), resulting in 76,554 positions in total. The whole genome alignment was analyzed using IQ-TREE version 1.6.6 (Nguyen et al. Citation2014) under the TIM3 + F + R3 model. The tree topology was verified under both 1000 bootstrap and 1000 replicates of SH-aLRT test. Phylogenetic analysis revealed that the genera Amentotaxus and Torreya form a sister group to the other genera of Taxaceae ().
Acknowledgements
We are grateful to Lei Cai for collecting the samples.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Bi G, Mao Y, Xing Q, Cao M. 2018. HomBlocks: a multiple-alignment construction pipeline for organelle phylogenomics based on locally collinear block searching. Genomics. 110:18–22.
- Dierckxsens N, Patrick M, Guillaume S. 2016. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:e18.
- Elpe C, Knopf P, Stützel T, Christian S. 2018. Diversity and evolution of leaf anatomical characters in Taxaceae s.l.—fluorescence microscopy reveals new delimitating characters. J Plant Res. 131:125–141.
- Huang DI, Cronk Q. 2015. Plann: a command-line application for annotating plastome sequences. Appl Plant Sci. 3:apps.1500026.
- Nguyen LT, Schmidt HA, Haeseler A, Minh BQ. 2014. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32:268–274.
- Nystedt B, Street NR, Wetterbom A, Zuccolo A, Lin Y-C, Scofield DG, Vezzi F, Delhomme N, Giacomello S, Alexeyenko A, et al. 2013. The Norway spruce genome sequence and conifer genome evolution. Nature. 497:579–584.
- Sun WB. 2013. Conserving Plant Species with Extremely Small Populations (PSESP) in Yunnan: a practice and exploration. Kunming: Yunnan Science and Technology Press; p. 11–14.
- Thomas P, Gao L, Phan KL. 2017. Amentotaxus yunnanensis. The IUCN Red List of Threatened Species 2017. e.T118076066A96812158. http://dx.doi.org/10.2305/IUCN.UK.2017-2.RLTS.T118076066A96812158.en. Downloaded on 24 January 2019.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.