Abstract
Isotomurus maculatus (Collembola, Isotomidae) is a springtail with a large distribution. This species has been introduced to the sub-Antarctic Prince Edward Islands. In this study, the mitogenome of I. maculatus was reconstructed. The total length of the mitochondrial genome is 15,263 bp and comprises 13 protein-coding genes, 22 tRNAs, and two rRNAs. The nucleotide composition is 34.4% adenine, 29.2% thymine, 15.1% guanine, and 21.3% cytosine. A Bayesian phylogenetic tree indicates that I. maculatus clusters as the sister taxon to the genus Cryptopygus, with high statistical support.
Collembola (springtails) are ubiquitous arthropods with representatives in most terrestrial ecosystems (Carapelli et al. Citation1995). Springtails are important in ecological systems due to their role in nutrient cycling and as bioindicators of ecosystem health (Rusek Citation1998; Filser Citation2002; Seastedt Citation1984). The geographical distribution of species tend to be dynamic and varies as a result of climatic changes or habitat suitability (Hill et al. Citation1999; Root et al. Citation2003). Species belonging to the genus Isotomurus have successfully colonized a wide range of habitats and have tentatively been described from sub-Antarctic oceanic islands. On Marion Island (one of two islands belonging to the Prince Edward Islands), Isotomurus cf. palustris have colonized the entire island, in spite of very low mitochondrial variation at the cytochrome oxidase subunit I gene (Myburgh et al. Citation2007). This possibly suggests that successful colonization involved only a few founders. This putative species was subsequently classified as Isotomurus maculatus (Greenslade Citation2010) based on morphological differences, notably the pigmentation pattern on the dorsal side of the animals (Carapelli et al. Citation2001; Greenslade Citation2010).
Isotomurus maculatus is a hemiedaphic springtail (lives partly within the soil and partly on the surface; Carapelli et al. Citation2001) and is a widespread species (Fjellberg Citation2007; Liu et al. Citation2012; Greenslade et al. Citation2013). Here, we report the annotated mitogenome of I. maculatus. This information, when taken with data available for other species (Torricelli et al. Citation2010; Jagatap et al. Citation2019; Monsanto et al. Citation2019) can be used to understand whether a selective adaptive advantage contributes to its success as a widespread species.
Genomic DNA of 16 specimens from the Prince Edward Islands (South African territory) was extracted using the DNeasy Blood and Tissue kit (Qiagen, Germany). Since whole specimens were used for DNA extractions, another specimen from the same locality was deposited as a voucher specimen at the Iziko Museums of South Africa (Voucher No. SAM-ENW-C010312). Genomic libraries were prepared and indexed separately. The libraries were sequenced using the Illumina HiSeq4000 platform (Illumina, USA). The sequencing run produced a total of 66 GB of raw data. The mitogenome was assembled using NOVOPlasty (Dierckxsens et al. Citation2017) with default settings, except for the insert size that was changed to 350 bp. The reference seed was the COI gene of I. maculatus (accession number: DQ147558; Myburgh et al. Citation2007). The MITOS2 WebServer (Donath et al. Citationin press) was used to annotate the genome. The protein-coding gene (PCG) boundaries were adjusted using Exonerate v2.2 (Slater and Birney Citation2005) and the ribosomal RNA boundaries were re-validated in Infernal v1.1 (Nawrocki and Eddy Citation2013). Each PCG of I. maculatus and nine other arthropods (>68% similarity) was aligned independently. A Bayesian tree was constructed in BEAST v2.5.0 (Bouckaert et al. Citation2014) using the HKY substitution model (Hasegawa et al. Citation1985), and 10 million MCMC iterations with a 20% burnin. The tree was visualised in Figtree v.1.4.3 (Rambaut Citation2016) ().
Figure 1 . Bayesian phylogenetic tree of Isotomurus maculatus and nine other species (seven Collembola and two arthropods) with the nodes showing well supported posterior probabilities. The following species were used for the phylogenetic analysis: Isotomurus maculatus MK509021 (Collembola, Isotomidae), Cryptopygus antarcticus NC_010533 (Collembola, Isotomidae), Cryptopygus terranovus NC_037610 (Collembola, Isotomidae), Folsomia candida KU198392 (Collembola, Isotomidae), Folsomotoma octooculata KC862316 (Collembola, Isotomidae), Orchesella cincta NC_032283 (Collembola, Entomobryidae), Orchesella villosa EU016195 (Collembola, Entomobryidae), Gomphiocephalus hodgsoni AY191995 (Collembola, Hypogastruridae), Anopheles stephensi KT899888 (Diptera, Culicidae), Simulium maculatum NC_040120 (Diptera: Simuliidae).
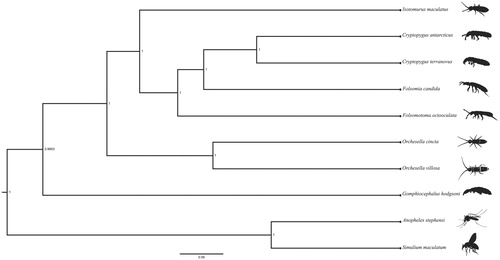
The total length of the circularised mitochondrial genome is 15,263 bp. Typical mitogenomic features (13 PCGs, 22 tRNAs, and two rRNAs) were annotated. The nucleotide composition of the genome is 34.4% adenine, 29.2% thymine, 15.1% guanine, and 21.3% cytosine. Based on available data, Isotomurus maculatus clusters as the sister taxon to the genus Cryptopygus, with high statistical support. The significant divergence between Collembola and other members of the Insecta is also confirmed by our results (Luan et al. Citation2005; Gao et al. Citation2008).
Acknowledgements
We thank the Department of Environmental Affairs (South African National Antarctic Programme) who allowed us to collect specimens (permit number 1/2013) and provided logistical support for the voyage. We acknowledge the Centre for High Performance Computing (CHPC) in Cape Town and IT service (University of Johannesburg) for bioinformatics support.
Disclosure statement
The authors report no conflict of interest, and are responsible for the content and writing of this article.
Additional information
Funding
References
- Bouckaert R, Heled J, Kühnert D, Vaughan T, Wu C-H, Xie D, Suchard MA, Rambaut A, Drummond AJ. 2014. BEAST 2: a software platform for Bayesian evolutionary analysis. PLoS Comput Biol. 10:e1003537.
- Carapelli A, Fanciulli PP, Frati F, Dallai R. 1995. The use of genetic markers for the diagnosis of sibling species in the genus Isotomurus (Insecta, Collembola). Italian J Zool. 62:71–76.
- Carapelli A, Frati F, Fanciulli PP, Dallai R. 2001. Taxonomic revision of 14 southwestern European species of Isotomurus (Collembola, Isotomidae), with description of four new species and the designation of the neotype for I. palustris. Zool Scr. 3:115–143.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:1–9.
- Donath A, Juhling F, Al-Arab M, Stadler PF, Middendorf M, Bernt M. in press. Improved annotation of protein-coding genes boundaries in metazoan mitochondrial genomes. Nucleic Acids Res. 1–10.
- Filser J. 2002. The role of Collembola in carbon and nitrogen cycling in soil. Pedobiologia. 46:234–245.
- Fjellberg A. 2007. Fauna Entomologica Scandinavica Volume 42. The Collembola of Fennoscandia and Denmark. Part II: Entomobryomorpha and Symphypleona. Leiden (The Netherlands): Brill.
- Gao Y, Bu Y, Luan YX. 2008. Phylogenetic relationships of basal hexapods reconstructed from nearly complete 18S and 28S rRNA gene sequences. Zool Sci. 25:1139–1145.
- Greenslade P. 2010. Collembola fauna of the South Shetland Islands revisited. Antarct Sci. 22:233–242.
- Greenslade P, Boyer S, Wratten S. 2013. New records of springtails in New Zealand pasture: how well are our pastoral invertebrates known? New Zealand J Agric Res. 56:93–101. DOI:10.1080/00288233.2012.754774.
- Hasegawa M, Kishino H, Yano T. 1985. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J Mol Evol. 22:160–174.
- Hill JK, Thomas CD, Huntley B. 1999. Climate and habitat availability determine 20th century changes in a butterfly’s range margin. Proc. R. Soc. London, Ser. B. 266:1197–1206.
- Jagatap H, Monsanto DM, Jansen van Vuuren B, Janion-Scheepers C, Sekar S, Teske PR, Emami-Khoyi A. 2019. The complete mitogenome of the springtail Tullbergia bisetosa: a species with a unique biology. Mitochondrial DNA B. 4:1594–1596.
- Liu WPA, Janion C, Chown SL. 2012. Collembola diversity in the critically endangered Cape Flats Sand Fynbos and adjacent pine plantations. Pedobiologia. 55:203–209.
- Luan YX, Mallatt JM, Xie RD, Yang YM, Yin WY. 2005. The phylogenetic positions of three basal-hexapod groups (Protura, Diplura, and Collembola) based on ribosomal RNA gene sequences. Mol Biol Evol. 22:1579–1592.
- Monsanto DM, Jansen van Vuuren B, Jagatap H, Jooste CM, Janion-Scheepers C, Teske PR, Emami-Khoyi A. 2019. The complete mitogenome of the springtail Cryptopygus antarcticus travei provides evidence for speciation in the Sub-Antarctic region. Mitochondrial DNA B. 4:1195–1197.
- Myburgh M, Chown SL, Daniels SR, Jansen van Vuuren B. 2007. Population structure, propagule pressure, and conservation biogeography in the sub-Antarctic: lessons from indigenous and invasive springtails. Divers Distrib. 13:143–154.
- Nawrocki EP, Eddy SR. 2013. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics. 29:2933–2935.
- Rambaut A. 2016. Figtree 1.4.3. Available from: http://tree.bio.ed.ac.uk/software/figtree/ [accessed 2019 February 5].
- Root TL, Price JT, Hall KR, Schneider SH, Rosenzweig C, Pounds JA. 2003. Fingerprints of global warming on wild animals and plants. Nature. 421:57–60.
- Rusek J. 1998. Biodiversity of Collembola and their functional role in the ecosystem. Biodivers Conserv. 7:1207–1219.
- Seastedt T. 1984. The role of microarthropods in decomposition and mineralization processes. Ann Rev Entomol. 29:25–46.
- Slater GS, Birney E. 2005. Automated generation of heuristics for biological sequence comparison. BMC Bioinform. 6:31–42.
- Torricelli G, Carapelli A, Convey P, Nardi F, Boore JL, Frati F. 2010. High divergence across the whole mitochondrial genome in the “pan-Antarctic” springtail Friesea grisea: evidence for cryptic species? Gene. 449:30–40.
