Abstract
The whole chloroplast genome sequence of Populus afghanica is characterized in this study. The size of cp genome of P. afghanica is 155,975 bp, containing a large single-copy region (LSC) of 84,223 bp, a small single-copy region (SSC) of 16,504 bp, and a pair of identical inverted repeat regions (IRa, b) of 27,624 bp. The overall GC content of the chloroplast genome is 36.8%. One hundred twenty-eight genes were annotated, including 81 protein-coding genes, 38 tRNA genes, and eight rRNA genes. The maximum-likelihood phylogenetic analysis with the reported chloroplast genomes showed that P. afghanica is sister to Populus lasiocarpa Oliv., implied a hybridization orign.
Introduction
Populus afghanica is a member of the section Aigeiros in genus Populus, its morphological feature is characterized as grayish bark, darker at base of trunk; wide crown and medium sized. Populus afghanica mainly distributed in the south of Xinjiang Province (China) and Pakistan, Afghanistan, Iran and the South of Dissolution of the Soviet union, the growing elevation above 1400 m (Wang and Fang Citation1984). Chloroplast (cp) genomes have the great taxonomic value because of their highly conserved structures with regard to gene content and gene order (Ravi et al. Citation2008; Wang et al. Citation2010; Wu and Ge Citation2012). There are extensive interspecific hybridization events among species in Populus. For example, P. afghanica belonging to the section Aigeiros, shared a common maternal ancestor with P. lasiocarpa of section Leucoides in the phylogenetic analysis (Wang et al. Citation2014). In this study, we reported the complete cp genome of P. afghanica, which can be useful in reconstructing high-resolution phylogenetic relationship and estimating the divergence times in the genus.
The fresh leaves of P. afghanica were collected from Xinjiang (39°28N, 75°59E, Altitude 1425 m), China. The voucher specimen deposited in the herbarium of Research Institute of Forestry, CAF. Total genomic DNA was extracted with a modified CTAB method (Doyle and Doyle Citation1987). Paired-end (PE) reads were constructed according to the Illumina library preparation protocol and sequenced on an Illumina HiSeq 2000 platform (Illumina, San Diego, CA). Then, we used the MITObim 1.8 to assemble the whole cp genome of P. afghanica (University of Oslo, Oslo, Norway; Kaiseraugst, Switzerland) (Hahn et al. Citation2013). The cp genome of P. lasiocarpa (GenBank accession: NC_036040.1) was used as the reference. The complete cp genome of P. afghanica was annotated in DOGMA (http://dogma.ccbb.utexas.edu/) (Wyman et al. Citation2004), and then corrected with Sequin 15.50 and Geneious 8.0.2 (Kearse et al. Citation2012).
After assembly, the total length of the cp genome of P. afghanica (GenBank accession MK294735) is 155,975 bp and is composed of LSC region of 84,223 bp, two IR copies of 27,624 bp, and SSC region of 16,504 bp. The overall GC contents of the cp genome were 36.8% and that of the LSC, SSC, and IR regions were 35%, 31%, and 42%, respectively. The proportion of coding sequences including all RNA regions is 36.7%. The cp genome includes 128 genes, including 81 protein-coding genes (75 protein-coding gene species), 39 tRNA genes (32 tRNA species), and eight rRNA genes (four rRNA species). Sixteen genes contain introns, and three of these genes, clpP, rps12, and ycf3, exhibit two introns. A total of 18 genes were duplicated in the inverted repeat regions including seven tRNAs, seven protein genes, and four rRNAs. Also, partial of ycf1 duplicated in IR region is formed as one pseudogene.
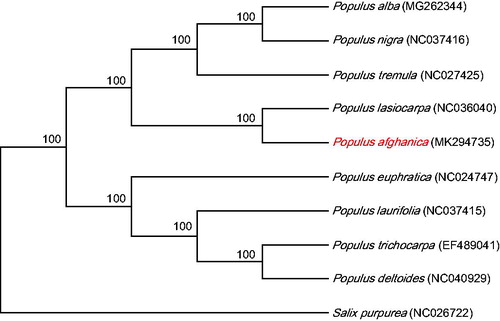
The maximum likelihood phylogenetic tree was constructed based on nine related species of Populus and one species of Salicaceae (Salix) using MEGA 7.0 (Kumar et al. Citation2016) from alignments created using the MAFFT (Katoh and Standley Citation2013). Consistent with previous studies, P. afghanica was closely grouped with P. lasiocarpa of section Leucoides (). Given the fact that P. afghanica belonged to the section Aigeiros according to the morphological analysis (Wang and Fang Citation1984), hybridization event should be involved during its speciation process. We reported the complete chloroplast genome of P. afghanica, to provide genomic and genetic sources for understanding more about the phylogenetic relationship of Populus.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads-a baiting and iterative mapping approach. Nucleic Acids Res. 41:e129.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- Ravi V, Khurana JP, Tyagi AK, Khurana P. 2008. An update on chloroplast genomes. Plant Syst Evol. 271:101–122.
- Wang C, Fang CF. 1984. Salicaceae. Flora of China. 20:1–403.
- Wang L, Qi XP, Xiang QP, Heinrichs J, Schneider H, Zhang XC. 2010. Phylogeny of the paleotropical fern genus Lepisorus (Polypodiaceae, Polypodiopsida) inferred from four chloroplast genome regions. Mol Phylogenet Evol. 54:211–225.
- Wang ZS, Du SH, Dayanandan S, Wang DS, Zeng YF, Zhang JG. 2014. Phylogeny reconstruction and hybrid analysis of Populus (Salicaceae) based on nucleotide sequences of multiple single-copy nuclear genes and plastid fragments. PLoS One. 9:e103645.
- Wu ZQ, Ge S. 2012. The phylogeny of the BEP clade in grasses revisited: evidence from the whole-genome sequences of chloroplasts. Mol Phylogenet Evol. 62:573–578.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.