Abstract
Ulva prolifera is the dominant species of green macroalgal blooms in the Yellow Sea, China. In this study, we sequenced and annotated the complete chloroplast genome of U. prolifera (GenBank accession number: KX342867). The genome of circular chromosomes consists of 93,066 including 66 protein-encoding genes, 26 tRNA genes, and 3 rRNA genes. Compared with the 16 species from Chlorophyta, it has eight genes in expansion and 15 genes in shrink. Phylogenetic analysis shows positioned U. prolifera with U. linza, while Ulva sp.UNA00071828 is closely allied with Ulva fasciata.
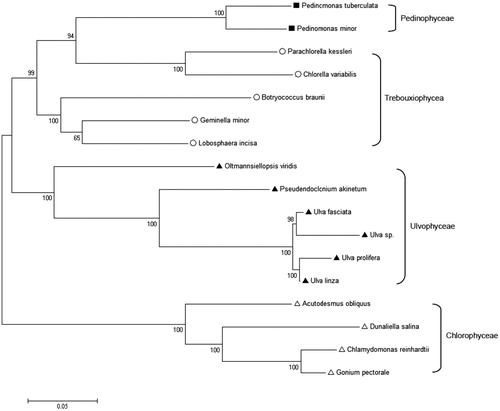
Green tide algae in Yellow Sea of China are mainly caused by the explosive growth of large planktonic green algae in shallow sea and intertidal area, wherein U. prolifera is considered as the dominant species floating to Qingdao (Han et al. Citation2013). Compared with the mitogenome, chloroplast genome contains more protein-coding genes, which plays an important role in cytoplasmic inheritance, plant phylogeny, DNA barcoding, and genetic diversity(Cai et al. Citation2017; Cai et al. Citation2018; Cremen et al. Citation2018). Thousands of chloroplast genomes have been published according to NCBI. However, the chloroplast genome of U. prolifera has never been reported before. Here, we sequenced and annotated it to gain insight into the mechanism of the green tides. Ulva prolifera was collected from the coast of Rudong sea area and cultured in laboratory with VSE medium at 20 °C under a light intensity of 100 μmolm−2 s−1 (Zhou et al. Citation2016a, Citation2016b). The complete chloroplast genome of Ulva prolifera is 93,066 bp in length, which is a double-chain closed loop missing reverse repeating area (Melton et al. Citation2015) (GenBank accession number: KX342867). The overall base composition of Ulva prolifera chloroplast genome is A (38.02%), T (37.02%), C (12.33%), G (12.45%), similar to most of the green algae in Chlorophyta. It contains 66 protein-encoding genes, 26 tRNA genes, and 3 rRNA genes. Compared with the 16 species from Chlorophyta (Leliaert and Lopez-Bautista Citation2015), it has eight genes in expansion (accD, chlI, ftsH, infA, psaI, rpl12, rpl19, ycf12) and 15 genes in shrink (chlB, chlL, chlN, cysA, cysT, minD, petL, psaM, psbT, rpl32, tilS, ycf20, ycf47, ycf62, psaK). In the phylogenetic tree constructed by genes from the chloroplast genome, which positions U. prolifera with U. linza, Ulva sp.UNA00071828 is closely allied with Ulva fasciata ().
Figure 1. The phylogenetic tree based on 51 genes of 17 Chlorophyta algaes. Ulva sp. UNA00071828 KP720616.1; Ulva fasciata NC_029040.1; Oltmannsiellopsis viridis NC_008099.1; Pseudendoclonium akinetum NC_008114.1; Chlamydomonas reinhardtii NC_005353.1; Dunaliella salina NC_016732.1; Gonium pectoral NC_020438.1; scenedesmus obliquus NC_008101.1; Pedinomonas minor NC_016733; Pedinomonas tuberculata NC_025530; Chlorella variabilis KJ718922; Parachlorella kessleri NC_012978; Botryococcus braunii NC_025545; Geminella minor NC_025544; Lobosphaera incise NC_025533.1; Ulva linza NC_030312.1.
Disclosure statement
We declare that we have no competing interests as defined by Nature Publishing Group or other interests that might be perceived to influence the results and discussion reported in this paper.
Additional information
Funding
References
- Cai C, Wang L, Jiang T, Zhou L, He P, Jiao B. 2018. The complete mitochondrial genomes of green tide algae Ulva flexuosa (Ulvophyceae, Chlorophyta).Conservation Genet Resour. 10:415–418.
- Cai C, Wang L, Zhou L, He P, Jiao B. 2017. Complete chloroplast genome of green tide algae Ulva flexuosa (Ulvophyceae, Chlorophyta) with comparative analysis. PLoS One. 12:e0184196.
- Cremen MCM, Leliaert F, Marcelino VR, Verbruggen H. 2018. Large diversity of non-standard genes and dynamic evolution of chloroplast genomes in siphonous green algae (Bryopsidales, Chlorophyta). Genome Biol Evol. 10:1048–1061.
- Han W, Chen L-P, Zhang J-H, Tian X-L, Hua L, He Q, Huo Y-Z, Yu K-F, Shi D-J, Ma J-H, et al. 2013. Seasonal variation of dominant free-floating and attached Ulva species in Rudong coastal area, China. Harmful Algae. 28:46–54.
- Leliaert F, Lopez-Bautista JM. 2015. The chloroplast genomes of Bryopsis plumosa and Tydemania expeditiones (Bryopsidales, Chlorophyta): compact genomes and genes of bacterial origin. BMC Genomics. 16:20.
- Melton JT, III, Leliaert F, Tronholm A, Lopez-Bautista JM. 2015. The complete chloroplast and mitochondrial genomes of the green macroalga Ulva sp UNA00071828 (Ulvophyceae, Chlorophyta). PLoS One. 10:e0121020.
- Zhou L, Wang L, Zhang J, Cai C, He P. 2016a. Complete mitochondrial genome of Ulva linza, one of the causal species of green macroalgal blooms in Yellow Sea, China. Mitochondr DNA B. 1:31–33.
- Zhou L, Wang L, Zhang J, Cai C, He P. 2016b. Complete mitochondrial genome of Ulva prolifera, the dominant species of green macroalgal blooms in Yellow Sea, China. Mitochondr DNA B. 1:76–78.
