AbSTRACT
Aporrectodea rosea is a cosmopolitan earthworm species widespread in agricultural soils throughout the world. In this paper, we present its complete mitochondrial genome, which is a circular molecule 15,086 bp long and has gene content and arrangement typical for earthworm mtDNA.
Despite the huge ecological importance of the Lumbricidae family (Hendrix et al. Citation2008), only one lumbricid mitochondrial genome was published for Lumbricus terrestris (Boore and Brown Citation1995). Mitochondrial genomes of representatives of other earthworm families from subtropical regions were also sequenced (Zhang et al. Citation2015; Wang et al. Citation2015; Zhang, Jiang, et al. Citation2016a, Citation2016b; Zhang, Sechi, et al. Citation2016).
Aporrectodea rosea is a widespread cosmopolitan earthworm found both in natural and agricultural habitats throughout the temperate zone (Vsevolodova-Perel Citation1997). This species is known for its high genetic diversity; the species is divided into Mediterranean and Eurosiberian lineages, each of which contains multiple genetic lineages that differ by 10–15% nucleotide substitutions in mtDNA (King et al. Citation2008; Fernández et al. Citation2016).
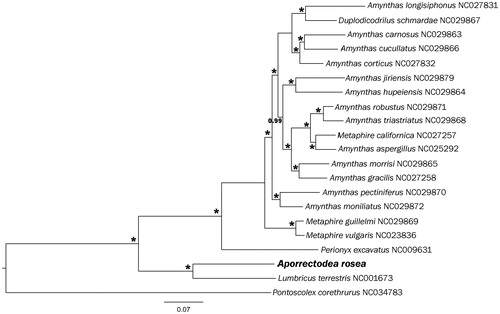
Aporrectodea rosea individuals were collected near the Vodnik station, to the north of Omsk, Russia (approximately N55.05 E73.40). Several ethanol-fixed individuals were identified morphologically. Total RNA was extracted from a typical specimen by Trizol; DNA was extracted in parallel, and by sequencing the coi barcode we confirmed that it belonged to an A. rosea L4 individual. Genomic DNA of the studied specimen (R41_Arosea) and ethanol-fixed specimens from the same population are stored in the collection of ICiG SB RAS in Novosibirsk. TruSeq Stranded mRNA kit (Illumina) was used to isolate and reverse transcribe poly-A mRNA. Single-end 150 bp reads were obtained on Illumina NextSeq 550 machine using a NextSeq 500 Mid Output Kit (Illumina). RNA reads were assembled using Trinity v.2.4.0 (Grabherr et al. Citation2011). Search for mitochondrial sequences was performed using the blastn, blastp, and blastx algorithms (Boratyn et al. Citation2013) with L. terrestris genome as a reference, which allowed us to recover A. rosea mtDNA sequence except for the control region. The latter could not be assembled from the RNAseq data due to repeats and was thus obtained by PCR using Herculase (Agilent Technologies). Phylogenetic analysis was performed using MrBayes v3.2.6 (Ronquist and Huelsenbeck Citation2003) based on the complete protein dataset of earthworm mitochondrial genomes ().
Figure 1. Bayesian tree of 13 concatenated protein sequences of earthworm mitochondrial genomes. Numbers near braches indicate Bayesian posterior probabilities; asterisk stands for 1.0.
The complete mtDNA sequence was annotated and deposited in GenBank under the MK573632 accession number. The complete mitochondrial genome of A. rosea L4 is 15,086 bp long and contains the typical gene complement of 13 protein-coding genes, 22 tRNAs, and 2 rRNAs. AT-content is 65.5%, and the coding strand is significantly enriched for both A (32%) and T (31%).
All genes are encoded by a single DNA strand and are thus probably transcribed as a single transcript. Compared to the L. terrestris mtDNA, the genome of A. rosea has a 56 bp deletion in the region between the cytb and tRNA-Trp genes. The control region located between the Arg- and His-tRNA genes is 512 bp long, which is about 130 bp longer than in L. terrestris, and contained a 130-bp long stretch of TA-microsatellite and a 22 bp long polyA tract. The nd4l and nd4 genes overlap by seven bp. All protein-coding genes start with ATG. Stop-codons are represented by TAA (for coii, nd6, nd5, and nd4l), or by TA or T (the rest of the genes), believed to be expanded to TAA by polyadenylation.
Phylogenetic analysis based on the 13 protein-coding genes () revealed that A. rosea groups together with L. terrestris is another representative of the Lumbricidae family.
Acknowledgements
The authors are grateful to Dr. Elena Golovanova for the earthworm specimens. Illumina sequencing was performed in the Center for Genome Studies of the Institute of Cytology and Genetics SB RAS.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Boore JL, Brown WM. 1995. Complete sequence of the mitochondrial DNA of the annelid worm Lumbricus terrestris. Genetics. 141:305–319.
- Boratyn GM, Camacho C, Cooper PS, Coulouris G, Fong A, Ma N, Madden TL, Matten WT, McGinnis SD, Merezhuk Y, et al. 2013. BLAST: a more efficient report with usability improvements. Nucleic Acids Res. 41:W29–W33.
- Fernández R, Novo M, Marchan DF, Diaz Cosin DJ. 2016. Diversification patterns in cosmopolitan earthworms: similar mode but different tempo. Mol Phylogenet Evol. 94:701–708.
- Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, et al. 2011. Full-length transcriptome assembly from RNA-seq data without a reference genome. Nat Biotechnol. 29:644–652.
- Hendrix PF, Callaham MA, Drake JM, Huang C-Y, James SW, Snyder BA, Zhang W. 2008. Pandora’s box contained bait: the global problem of introduced earthworms. Annu Rev Ecol Evol Syst. 39:593–613.
- King RA, Tibble AL, Symondson WOC. 2008. Opening a can of worms: unprecedented sympatric cryptic diversity within British lumbricid earthworms. Mol Ecol. 17:4684–98.
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic interference under mixed models. Bioinformatics. 19:1572–1574.
- Vsevolodova-Perel TS. 1997. The earthworms of the fauna of Russia. Moscow: Nauka.
- Wang AR, Hong Y, Win TM, Kim I. 2015. Complete mitochondrial genome of the Burmese giant earthworm, Tonoscolex birmanicus (Clitellata: Megascolecidae). Mitochondrial DNA. 26:467–468.
- Zhang L, Jiang J, Dong Y, Qiu J. 2015. Complete mitochondrial genome of four pheretimoid earthworms (Clitellata: Oligochaeta) and their phylogenetic reconstruction. Gene. 574:308–316.
- Zhang L, Jiang J, Dong Y, Qiu J. 2016a. Complete mitochondrial genome of an Amynthas earthworm, Amynthas aspergillus (Oligochaeta: Megascolecidae). Mitochondrial DNA A. 27:1876–1877.
- Zhang L, Jiang J, Dong Y, Qiu J. 2016b. Complete mitochondrial genome of a Pheretimoid earthworm Metaphire vulgaris (Oligochaeta: Megascolecidae). Mitochondrial DNA A. 27:297–298.
- Zhang L, Sechi P, Yuan M, Jiang J, Dong Y, Qiu J. 2016. Fifteen new earthworm mitogenomes shed new light on phylogeny within the Pheretima complex. Sci Rep. 6:20096.
