Abstract
Here, we report the first complete mitochondrial genome of Gekko subpalmatus. The genome of Gekko subpalmatus is 17,105 bp in length and consists of 13 protein-coding genes, 22 transfer RNA genes, 2 ribosomal RNA genes, and 1 control region. The phylogenetic tree reveals that G. subpalmatus is a sister group to G. hokouensis and shows (((((G. subpalmatus, G. hokouensis), G. swinhonis), G. japonicus), G. chinensis), G. gecko) with strong support. This study provides more molecular data for G. subpalmatus and lays the foundation for future protection.
The webbed-toed gecko, Gekko subpalmatus, belongs to the Gecko family (Gekkonidae) (Luu et al. Citation2014). This species is endemic to China and distributed in southern China, including the Hong Kong and Hainan (Brown et al. Citation2012). Due to the expansion of human activities and habitat destruction, G. subpalmatus had been listed in the IUCN Red List (NT) in the 2013 and had received more attention for the biodiversity conservation. However, to date, there have been very few reports on this species, based on the short-fragment mitochondrial genes that are not comprehensive enough to understand the information for G. subpalmatus (Rato and Harris Citation2008), whether in phylogenetic analysis or evolutionary biology. This study provides the first complete mitochondrial genome of G. subpalmatus and helps to further understand the information and phylogeny of G. subpalmatus.
The sample of G. subpalmatus was collected from Chengdu (N30°70′70″, E103°86′63″), Sichuan Province, China in 2018, and tail tip tissue was stored in −80 °C ultra-low temperature freezer. Total DNA was isolated using Ezup-pillar Genomic DNA Extraction Kit (Sangon, Shanghai, China). A DNA sample was sent to the Personal Biotechnology Co, Ltd (Shanghai, China) to obtain a complete mitochondrial genome via next-generation sequencing methods (Metzker Citation2010).
The length of the G. subpalmatus mitochondrial genome sequence is 17,105 bp (GenBank accession number MK680534). The base composition is 31.7% A, 27.1% T, 27.0% C, and 14.2% G in G. subpalmatus, which is with positive AT-skew (0.08) and negative GC-skew (−0.31). It contains 13 protein-coding genes, 22 transfer RNA genes, 2 ribosomal RNA genes, and 1 control region. Eight tRNA genes and ND6 are encoded on the L-strand (–), and the remaining genes are encoded on the H-strand (+). The length of tRNAs range from 64 to 75 bp and the 12S rRNA and 16S rRNA are 961 and 1560 bp in size, respectively. In addition, the control region is 1739 bp in size, which is located between the tRNA-Pro and tRNA-Phe. The arrangement and transcriptional direction of genes are consistent with previous studies in Gekkonidae (Zhou et al. Citation2006) and consistent with most typical vertebrate mitochondrial genomes (Anderson et al. Citation1982).
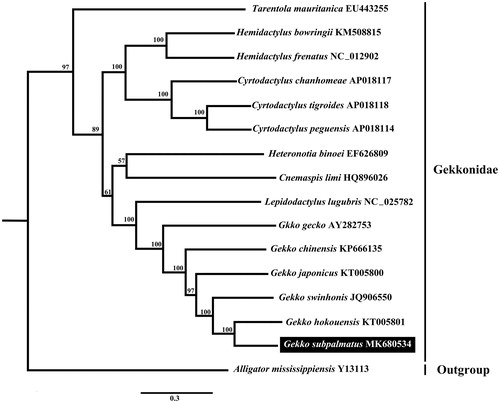
Fifteen Gekkonidae mitochondrial genomes including the new one obtained in this study were used for phylogenetic analyses and the Alligator mississippiensis was selected as an outgroup. Phylogenetic trees were constructed by maximum likelihood (ML) methods using Phyml 3.0 (with 1000 bootstrap replicates) (Guindon et al. Citation2010), the best-fitting nucleotide substitution model (GTR + I+G) was selected by model generator (Keane et al. Citation2006). The phylogenetic tree () revealed that G. subpalmatus was a sister group to G. hokouensis and showed (((((G. subpalmatus, G. hokouensis), G. swinhonis), G. japonicus), G. chinensis), G. gecko) with strong support. In addition, the phylogenetic relationship of Gekkonidae in this study was consistent with the previously described phylogeny using partial sequences of mitochondrial and nuclear genes (Heinicke et al. Citation2012).
Acknowledgements
We thank Ming Zhou in our lab for his help in data analysis.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Anderson S, Bruijn MH, De, Coulson AR, Eperon IC, Sanger F, Young IG. 1982. Complete sequence of bovine mitochondrial DNA. Conserved features of the mammalian mitochondrial genome. J Mol Biol. 156:683–717.
- Brown RM, Siler CD, Das I, Min Y. 2012. Testing the phylogenetic affinities of Southeast Asia's rarest geckos: flap-legged geckos (Luperosaurus), Flying geckos (Ptychozoon) and their relationship to the pan-Asian genus Gekko. Mol Phylogenet Evol. 63:915–921.
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 59:307–321.
- Heinicke MP, Greenbaum E, Jackman TR, Bauer AM. 2012. Evolution of gliding in Southeast Asian geckos and other vertebrates is temporally congruent with dipterocarp forest development. Biol Lett. 8:994–997.
- Keane TM, Creevey CJ, Pentony MM, Naughton TJ, Mclnerney JO. 2006. Assessment of methods for amino acid matrix selection and their use on empirical data shows that ad hoc assumptions for choice of matrix are not justified. BMC Evolutionary Biology. 6:29.
- Luu VQ, Calame T, Nguyen TQ, Le MD, Bonkowski M, Ziegler T. 2014. A new species of the Gekko japonicus group (Squamata: Gekkonidae) from central Laos. Zootaxa. 3895:73–88.
- Metzker ML. 2010. Sequencing technologies – the next generation. Nature Reviews Genetics. 11:31–46.
- Rato C, Harris D. 2008. Genetic variation within Saurodactylus and its phylogenetic relationships within the Gekkonoidea estimated from mitochondrial and nuclear DNA sequences. Amphibia-Reptilia. 29:25–34.
- Zhou K, Li H, Han D, Bauer AM, Feng J. 2006. The complete mitochondrial genome of Gekko gecko (Reptilia: Gekkonidae) and support for the monophyly of Sauria including Amphisbaenia. Mol Phylogenet Evol. 40:887–892.