Abstract
The complete mitogenome (106,827 bp) of the Antarctic moss Bartramia patens Brid. (Bartramiaceae) was analyzed. It consists of 40 protein-coding genes, 3 ribosomal RNAs, and 24 transfer RNAs. The phylogenetic tree based on the combined amino acids sequences of 32 mitochondrial genes showed B. patens to have a sister relationship with B. pomiformis. The size of the mitochondrial genome of B. patens was 629 bp larger than that of B. pomiformis due to the expansion of the intronic region. This phenomenon is very characteristic and requires more comparative studies with the mitogenomes of other bryophytes.
Keywords:
Antarctic moss is valuable for research in physiological, biochemical, and evolutionary aspects, but the mitogenomes have rarely been reported in Antarctic mosses (Yoon et al. Citation2016). Several species of Bartramiaceae (Bartramiales) grow in diverse habitats, and this family is considered as one of the poikilohydric groups (Vitt Citation1984). Initial studies of mitogenomes in genus Bartramia have been performed in B. pomiformis distributed in North America (Liu et al. Citation2014). Here, we report the mitogenome of Antarctic B. patens mainly distributed in Southern Hemisphere including Australia, New Zealand, South America and Antarctica (Matteri Citation1984, Citation1985; Streimann and Curnow Citation1989).
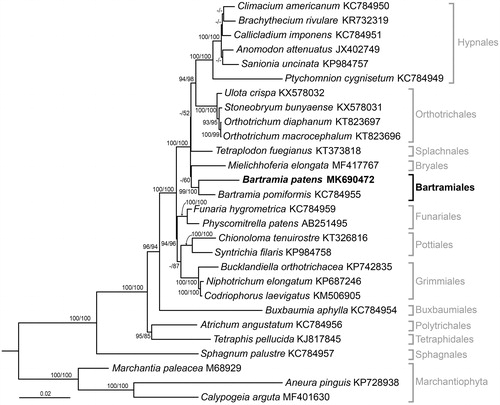
Moss samples were collected from a 3 × 3 cm patch of Bartramia patens population in the vicinity of the Korean King Sejong Antarctic Station (62°14′29″S; 58°44′18″W), on the Barton Peninsula of King George Island, in January 2014. The specimen of B. patens was deposited into KOPRI Herbarium (KOPRI-MO00225, accessible to https://kvh.kopri.re.kr). Total genomic DNA was prepared from two growth patches of B. patens using TruSeq DNA sample preparation kits (Illumina, San Diego, CA, USA) and sequenced by Illumina MiSeq in a paired-end mode (2 × 300 bp) (Illumina, USA). A total of 90,597 reads were used for de novo assembly of CLC Genomics Workbench V7.5 (CLC bio, Aarhus, Denmark). BLAST search, annotation and RNA verification were performed as previously described (Yoon et al. Citation2016). A total of 106,827 bp of B. patens mitogenome was obtained (NCBI accession ID: MK690472). It consists of 40 protein-coding genes, 3 ribosomal RNAs (rRNAs), and 24 transfer RNAs (tRNAs). The mitogenome structure and gene order were congruent with those of other Bryophyta. The phylogenetic analyses of B. patens was carried out using MAFFT version 7 for amino acids sequences of 32 genes (8094 amino acids in length) derived from 25 Bryophytes and 3 Marchantiophyta species accessible from the public DB (Katoh and Standley Citation2013). Multiple alignments were performed in maximum likelihood (ML) tree inferred with JTT + G + I + F model (lnL = –42,822.6621) selected by the Bayesian Information Criterion (BIC) score in MEGA7 (Kumar et al. Citation2016). ML and neighbor-joining (NJ) trees were constructed in MEGA7, and the tree topology was drawn in the ML tree (). The bootstrap values of ML and NJ were presented on the branches (above 80 and 50, respectively). Bartramia patens formed a sister relationship with B. pomiformis phylogenetically. Bartramiales made a clade with Bryales, Splachnales, Orthotrichales, and Hypnales. Interestingly, the mitogenome size of B. patens was 629 bp larger than that of B. pomiformis, resulting from the expansion of 689 bp of the genic regions (60 bp of exonic, 629 bp of intronic) and reduction of 60 bp of the intergenic regions. The expansion of intronic region was often found in mitogenomes. Thus, future comparative studies of mitogenomes between B. patens and other species would give an insight on the evolution of bryophytes.
Figure 1. Maximum likelihood (ML) tree inferred from amino acids of 32 mitochondrial genes with JTT + G+I + F parameters in MEGA7. Bootstrap analyses were conducted in ML and NJ with 1000 replicates and it was showed on branches above 80 of ML and 50 of NJ tree, respectively. Bartramia patens has a sister relationship with B. pomiformis which is distributed in North America than in Southern Hemisphere.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- Liu Y, Rafael M, Goffinet B. 2014. 350 Million years of mitochondrial genome stasis in mosses, an early land plant lineage. Mol Biol Evol. 31:2586–2591.
- Matteri CM. 1984. Sinopsis de las especies andino-patagónicas, antárcticas y subantárcticas de los géneros Bartramia, Bartramidula y Conostomum. Darwiniana. 25:143–162.
- Matteri CM. 1985. Bryophyta, Musci. Bartramiaceae. Flora Criptogámica de Tierra Del Fuego. 14:62.
- Streimann H, Curnow J. 1989. Catalogue of mosses of Australia and its external erritories. Australian Flora Fauna. 10:1–479.
- Vitt DH. 1984. Classification of the Bryopsida. In: Schuster RM, editor. New manual of bryology. 2nd ed. Nichinan: Hattori Botanical Laboratory; p. 696–759.
- Yoon YJ, Kang Y, Kim MK, Lee J, Park H, Kim JH, Lee H. 2016. The complete mitochondrial genome of an Antarctic moss Syntrichia filaris (Müll. Hal.) RH Zander. Mitochondrial DNA A. 27:2779–2780.
