Abstract
Cinnamomum chago B.S. Sun & H.L. Zhao is an economically important wild woody oil plant of the genus Cinnamomum Trew belonging the family Lauraceae. To better determine its phylogenetic location with respect to the other Cinnamomum species, the complete plastid genome of C. chago was sequenced. The whole plastome is 152,753 bp in length, consisting of a pair of inverted repeat (IR) regions of 20,074 bp, one large single-copy (LSC) region of 93,722 bp, and one small single-copy (SSC) region of 18,883 bp. The overall GC content of the whole plastome is 39.2%. Further, maximum likelihood phylogenetic analyse was conducted using 12 complete plastomes of the Lauraceae, which support the close relationship between C. chago and C. verum.
Cinnamomum chago B.S. Sun & H.L. Zhao, a narrowly distributed species at high altitudes in Yunnan of SW China and North Myanmar, was assigned to the genus Cinnamomum Trew in the family Lauraceae (Sun and Zhao Citation1991; Dong et al. Citation2016). The fruits of C. chago are rich in fatty oils and thus represent important wild woody oil plants in the genus Cinnamomum Trew (Sun and Zhao Citation1991). For a better understanding of the relationships of C. chago and other Cinnamomum species, it is necessary to reconstruct a phylogenetic tree based on high-throughput sequencing approaches.
Fresh young leaves of C. chago in Yunlong County (Yunnan, China; Long. 99.9412 E, Lat. 25.5701 N, 2249 m) were picked for DNA extraction (Doyle and Dickson Citation1987). The voucher was deposited at the Biodiversity Research Group of Xishuangbanna Tropical Botanical Garden (Accession Number: XTBG-BRG-SY34994). The whole plastid genome was sequenced following Zhang et al. (Citation2016), and their 15 universal primer pairs were used to perform long-range PCR for next-generation sequencing. The contigs were aligned using the publicly available plastid genome of Sassafras tzumu (GenBank accession number MF939339) (Song et al. Citation2017) and annotated in Geneious 4.8.
The plastome of C. chago (LAU00078), with a length of 152,753, was 34 bp and 32 bp larger than that of C. bodinieri (152,719 bp, MH394,415) and C. camphora (152,721 bp, MH050970). It was also 7 bp and 13 bp smaller than that of C. parthenoxylon (152,760 bp, MH050971) and C. verum (152,766 bp, KY635878). The length of the inverted repeats (IRs), large single-copy (LSC), and small single-copy (SSC) regions of C. chago was 20,074 bp, 93,722 bp, and 18,883 bp, respectively. The overall G + C content is 39.2% (LSC, 37.9%; SSC, 33.9%; IR, 44.4%).
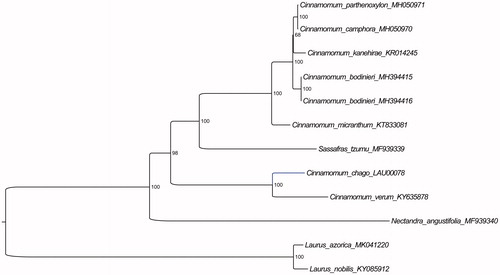
Furthermore, based on 15 published plastid genome sequences, we reconstructed a phylogenetic tree () to confirm the evolutionary relationship between C. chago and other species with published plastomes in Cinnamomum, with Laurus species as outgroup. Maximum likelihood (ML) phylogenetic analyses were performed based on GTR + F + I model in the iqtree version 1.6.7 program with 1000 bootstrap replicates (Nguyen et al. Citation2015). The ML phylogenetic tree with 47–100% bootstrap values at each node supported the fact that Cinnamomum species can be grouped into two clades, and that C. chago and C. verum were located in the same clade.
Data archiving statement
The plastome data of the C. chago will be submitted to Lauraceae Chloroplast Genome Database (LCGDB). Accession numbers are LAU00078.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Dong WJ, Zhang X, Yang GS, Yang L, Wang YH, Shen SK. 2016. Biological characteristics and conservation genetics of the narrowly distributed rare plant Cinnamomum chago (Lauraceae). Plant Divers. 38:247–252.
- Doyle JJ, Dickson EE. 1987. Preservation of plant samples for DNA restriction endonuclease analysis. Taxon. 36:715–722.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32:268–274.
- Song Y, Yu WB, Tan YH, Liu B, Yao X, Jin JJ, Padmanaba M, Yang JB, Corlett RT. 2017. Evolutionary comparisons of the chloroplast genome in Lauraceae and insights into loss events in the Magnoliids. Genome Biol Evol. 9:2354–2364.
- Sun BX, Zhao HL. 1991. A new species of Cinnamomum from Yunnan. J Yunnan Univ. 13:93–94.
- Zhang T, Zeng CX, Yang JB, Li HT, Li DZ. 2016. Fifteen novel universal primer pairs for sequencing whole chloroplast genomes and a primer pair for nuclear ribosomal DNAs. J Sytemat Evol. 54:219–227.