Abstract
In the present study, we reported the complete mitochondrial genome sequence of the medical spiny pipehorse Solegnathus spinosissimus. It was 16,530 bp in total length and contained 13 protein-coding genes, two rRNA genes, 22 tRNA genes, and a control region. The overall base composition of S. spinosissimus was 26.94%, 30.21%, 17.54%, and 25.31% for A, C, G, and T, respectively. The length of 12S and 16S rRNA were 938 bp and 1667 bp, respectively. The phylogenetic analysis showed that S. spinosissimus was most closely related to S. hardwickii, providing additional evidence to support the previous morphological classification of genus Solegnathus. The molecular data of S. spinosissimus could contribute to the genetic diversity and species identification study in Syngnathidae family.
The genus Solegnathus belongs to the family Syngnathidae, which is one of the important sources for traditional Chinese medicine (Asem et al. Citation2018). The medical spiny pipehorse Solegnathus spinosissimus (Günther, 1870) is one large species in the genus Solegnathus and often traded as adulteration of traditional Chinese medicinal pipefish (Gao et al. Citation2018). It mainly distributed in southwest-Pacific and preferred to live in areas with muddy bottoms at depths of 30–250 m (Dawson Citation1982; Martin-Smith and Vincent Citation2006; Forrest et al. Citation2008). Since the lack of information about the population size, trends in abundance and impacts of bycatch, S. spinosissimus has been listed in the IUCN as Data Deficient (Pollom Citation2017). The Complete mitochondrial genome provides valuable molecular data which would be useful for the phylogenetic analysis and further conservation strategy development (Chen et al. Citation2018; Zhu et al. Citation2018).
Here, we sequenced the complete mitochondrial genome of S. spinosissimus and constructed the phylogenetic to analysis relationships among Syngnathidae species. The specimen of S. spinosissimus was obtained from Panan herbal medicine market at Zhejiang Province and identified based on morphological characteristics including the extremely large body, snowflake pattern and trunk with spiny (Liu et al. Citation2018). The sample of S. spinosissimus DJ-02 was deposited in the collection center of Zhejiang Chinese Medical University. Total genomic DNA of S. spinosissimus was extracted from the tail muscle and the complete mtDNA was determined according to our previous reports (Ge et al. Citation2018; Fang et al. Citation2018; Lai et al. Citation2019). The complete mtDNA sequence of S.spinosissimus has been submitted to GenBank with the accession number of MK704428.
The complete genome of S. spinosissimus was 16,530 bp in length containing 13 PCGs, 22 tRNA genes, two rRNA genes, and one control region. The overall base compositions of S. spinosissimus were 26.94%, 30.21%, 17.54%, and 25.31% for A, C, G, and T, respectively. All PCGs started with the typical codon ATG except that COX1 started with GTG. The complete stop codon TAA was found in six genes, and the others stopped with a single T or TA. The length of 12S rRNA and 16S rRNA were 938 bp and 1667 bp, respectively. The 22 tRNA genes ranged from 67 to 75 bp in length. The control region was 917 bp in length locating between tRNA-Pro and tRNA-Phe.
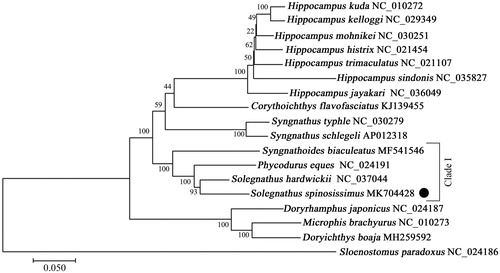
The phylogenetic relationships of S. spinosissimus and other Syngnathidae species were inferred utilizing maximum-likelihood (ML) based on the complete mitochondrial sequences. As showing in , S. spinosissimus and S. hardwickii clustered together with high statistical support, indicating a close genetic relationship. Furthermore, the genera Solegnathus, Phycodurus, and Syngnathoides consisted into one clade, suggesting species in genus Phycodurus could be applied as potential traditional Chinese medicine. Our results provide basic molecular data of S. spinosissimus, which would contribute to the phylogenetic analysis and species identification and conservation strategy in family Syngnathidae.
Figure 1. Phylogenetic tree showing the relationship between Solegnathus spinosissimus and 16 other Syngnathidae species based on the 13 PCGs sequences. The bootstrap value for the ML analysis based on 100 replicates was shown on each node. The Genbank accession number was listed following the species name.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Asem A, Xu Y-S, Wang P-Z, Li W. 2018. The complete mitochondrial genome of the pallid seahorse Solegnathus hardwickii (Actinopterygii; Syngnathiformes; Syngnathidae) obtained using next-generation sequencing. Mitochondrial DNA B. 3:923–924.
- Chen M, Zhu L, Chen J, Zhang G, Cheng R, Ge Y. 2018. The complete mitochondrial genome of the short snouted seahorse Hippocampus hippocampus, Linnaeus 1758 (Syngnathiformes: Syngnathidae) and its phylogenetic implications. Conservation Genet Resour. 10:783–787.
- Dawson CE. 1982. Synopsis of the Indo-Pacific genus Solegnathus (Pisces: Syngnathidae). Jpn J Ichthyol. 29:139–161.
- Fang Y, Zhu L, Chen M, Ge Y, Zhang G, Cheng R. 2018. Characterization of the complete mitochondrial genome of the medical pipefish Doryichthys boaja bleeker 1850. Mitochondrial DNA B. 3:881–883.
- Forrest RE, Scandol JP, Pitcher TJ. 2008. Towards ecosystem-based fishery management in New South Wales. Proceedings of the Experts and Data Workshop, December 8–10, 2003, Cronulla, Australia. Fish Cent Res Rep, p. 94.
- Gao L, Yin Y, Li J, Yuan Y, Jiang C, Gao W, Zhang X, Huang L. 2018. Identification of traditional Chinese medicinal pipefish and exclusion of common adulterants by multiplex PCR based on 12S sequences of specific alleles. Mitochondrial DNA A DNA Mapp Seq Anal. 29:340–346.
- Ge Y, Zhu L, Chen M, Zhang G, Huang Z, Cheng R. 2018. Complete mitochondrial genome sequence for the endangered Knysna seahorse Hippocampus capensis Boulenger 1900. Conservation Genet Resour. 10:461–465.
- Lai M, Sun S, Chen J, Lou Z, Qiu F, Zhang G, Cheng R. 2019. Complete mitochondrial genome sequence for the seahorse adulteration Hippocampus camelopardalis Bianconi 1854. Mitochondrial DNA B. 4:432–433.
- Liu F, Jin Y, Yuan Y, Qin W, Jiang C, Zhao Y, Huang L. 2018. Survey on origin of medicinal commodities of Syngnathus based on morphology and DNA sequencing identification. World Chin Med. 2:241–247.
- Martin-Smith KM, Vincent A. 2006. Exploitation and trade of Australian seahorses, pipehorses, sea dragons and pipefishes (Family Syngnathidae). Oryx. 40:141–151.
- Pollom R. 2017. Solegnathus spinosissimus. The IUCN Red List of Threatened Species 2017: e.T20318A67623085.
- Zhu L, Chen M, Cheng R, Ge W, Zhang G, Ge Y. 2018. Complete mitochondrial genome characterization of the alligator pipefish Syngnathoides biaculeatus and phylogenetic analysis of family Syngnathidae. Conserv Genet Resour. DOI:10.1007/s12686-018-1032-1.
