Abstract
The complete chloroplast (cp) genome of Rosa pricei Hayata has been characterized by reference-based assembly using Illumina paired-end data. The complete cp genome is 156,599 base pairs (bp) in length, containing a large single-copy region (LSC) of 85,732 bp, a small single-copy region (SSC) of 18,749 bp, which are separated by a pair of inverted repeat regions (IRs) of 26,059 bp, a total of 139 genes were predicted from the chloroplast genome, including 87 protein-coding genes, 39 tRNA genes, and eight rRNA genes (four rRNA species). Most genes occur as a single copy while 19 gene species are duplicated. Phylogenetic analysis revealed that R. pricei was resolved as sister to R. multiflora, R. lucieae, and R. maximowicziana.
Rosa pricei Hayata (Hayata Citation1915) is an endemic to Taiwan Island of China, distributed at altitudes of 600–2000 m. Its inflorescence has 3-9(-15) in terminal cyme and flowers white. The main distinction lies in the inflorescence and pubescent or glandular hairs on the phyllopodes or on the both surfaces of the sepals. In this study, we first reported the complete chloroplast genome of R. pricei.
A wild individual of R. pricei was sampled from its type locality in Central Range, Taiwan. The voucher specimen was deposited in the Herbarium of Chengdu Institute of Biology (CDBI: GXF15596). The total genomic DNA was extracted from leaves dried by silica gel using a modified CTAB method (Doyle and Doyle Citation1987) and sequenced based on the Illumina pair-end technology. The filtered reads were assembled using the program NOVOPlasty (Dierckxsens et al. Citation2017) with complete chloroplast genome of R. ordoata var.gigantea as the reference (GenBank accession No. KF753637). The assembled chloroplast genome was annotated using Plannv.1.1 (Huang and Cronk Citation2015), and the annotation result was modified using Geneiousv.10.2.3 (Kearse et al. Citation2012). The sequences were aligned using the MAFFT (Katoh and Standley Citation2013) using the published plastid genomes (excluding rose hybrids and cultivated species). Poorly aligned regions were trimmed using Gblocks v.0.91b (Castresana Citation2000) with the option ‘-t = c’ (the type of sequence was set to codons). The maximum-likelihood (ML) tree was constructed using RAxML v.8.2.11 (Stamatakis Citation2006) with 1000 bootstraps replicates to examine the phylogenetic position of R. pricei in Rosa genus ().
Figure 1. Phylogeny of chloroplast genome dataset, maximum-likelihood bootstrap support values are shown along the branches.
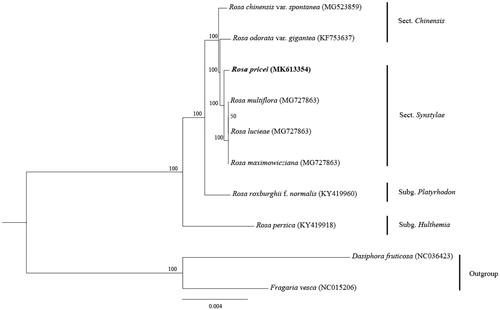
The complete chloroplast genome of R. pricei (Genbank accession No. MK613354) was a circular molecular genome with a size of 156,599 bp in length, which presented a typical quadripartite structure containing two inverted repeat (IR) regions of 26,059 bp separated by the large single-copy (LSC) region of 85,732 bp and small single-copy (SSC) region of 18,749 bp. The overall GC content was about 37.2%. The chloroplast genome consists of 139 genes including 87 protein coding genes, 39 tRNA genes, and eight rRNA genes (four rRNA species). Most genes occur as a single copy, while 19 gene species are duplicated. Our phylogenetic results showed that R. pricei was nested in the sect. Synstylae and resolved as sister to R. multiflora, R. lucieae, and R. maximowicziana. This complete chloroplast genome can be further studies for phylogenetic analyses of genus Rosa.
Disclosure statement
None of the co-authors has any conflict of interest to declare. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 17:540–552.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:e18.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Hayata B. 1915. Rosa Linn. Icon. Pl. Formosan. 5:58–60.
- Huang DI, Cronk Q. 2015. Plann: a command-line application for annotating plastome sequences. Appl Plant Sci. 3:1500026.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 22:2688–2690.
