Abstract
The complete mitochondrial genome of the tomato leafminer Tuta absoluta was determined in this study. It is 15,290 bp in length and consists of 13 protein-coding genes (PCGs), 22 tRNA genes, two rRNA genes, and a putative control region. All the 13 PCGs were initiated with typical ATN start codons, except for cox1 with the CGA. These PCGs were terminated by the typical TAA codon or the incomplete T– – codon. The phylogenetic relationships based on neighbour-joining method revealed that T. absoluta is clustered with six Gelechiidae species, which agree with the conventional taxonomy.
Keywords:
The tomato leafminer, Tuta absoluta (Meyrick) (Lepidoptera: Gelechiidae), is one of the most destructive pests in tomato production (Desneux et al. Citation2010, Citation2011). It is a native pest of South America but has spread to Africa, Europe, Asia, and Central America (Campos et al. Citation2017). Immediately after egg hatching, T. absoluta larvae bore tunnels into the leaves by feeding on the mesophyll (Desneux et al. Citation2011). In this study, adult individuals of T. absoluta were collected from Spain in 2009. The specimens were preserved in 95% ethanol and deposited in our laboratory (State Key Laboratory for Biology of Plant Diseases and Insect Pests, Chinese Academy of Agricultural Sciences).
The complete mitogenome of T. absoluta (GenBank accession number MK234840) is 15,290 bp in length, and it harbours the typical set of 37 mitochondrial genes, including 13 protein-coding genes (PCGs), 22 transfer RNA genes (tRNAs), two ribosomal RNA genes (rrnL and rrnS), and a putative control region (Boore Citation1999). Twenty-three genes were encoded on the heavy strand (H-strand), including nine PCGs and 14 tRNAs, whereas the others were located on the light strand (L-strand). The composition of this mitogenome is highly asymmetric (39.25% A, 12.05% C, 8.16% G, and 40.54% T), with an overall A + T content of 79.79%. The AT-skew and GC-skew of this mitogenome were −0.016 and −0.192, respectively. The gene composition and structure of T. absoluta were identical with other Gelechiidae species (Yuan et al. Citation2019).
There were four intergenic spacer regions comprising a total length of 19 bp and the largest spacer sequence of 8 bp resided between trnW and trnC. Gene overlaps were found at 17 gene junctions and involved a total of 198 bp, the longest 69 bp overlapping located between nad3 and trnA. The 13 PCGs were initiated at typical ATN start codons, except for cox1 with the CGA. All PCGs were terminated with the typical TAA codon except for cox1, cox2, nad1, and nad5, which terminated with a single T– – residue. The length of tRNAs varied between 64 bp (trnR) and 74 bp (trnS2), comprising a total length of 1474 bp. The 22 tRNA genes have cloverleaf-shaped structure and trnS1 gene lacks the dihydrouridine arm, which is similar to those reported in other animal mitogenomes (Wolstenholme Citation1992). The rrnL gene is located between trnL1 and trnV, whereas the rrnS gene between trnV and the control region. The length of rrnLand rrnS is 1344 bp and 776 bp, and their A + T content is 83.33% and 85.44%, respectively. The control region is located between rrnS and trnM and has a remarkably high A + T content (91.88%).
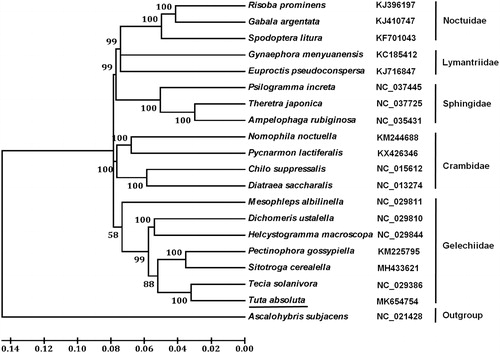
Based on the concatenated amino acid sequences of 13 PCGs, the neighbour-joining method was used to construct the phylogenetic relationship of T. absoluta with other moths. The result shows that T. absoluta is clustered with other six Gelechiidae species (), which agree with the conventional taxonomy.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Boore JL. 1999. Survey and summary: animal mitochondrial genomes. Nucleic Acids Res. 27:1767–1780.
- Campos MR, Biondi A, Adiga A, Guedes RNC, Desneux N. 2017. From the western palaearctic region to beyond: Tuta absoluta 10 years after invading Europe. J Pest Sci. 90:787–796.
- Desneux N, Wajnberg E, Wyckhuys KAG, Burgio G, Arpaia S, Narváez-Vasquez CA, González-Cabrera J, Ruescas DC, Tabone E, Frandon J, et al. 2010. Biological invasion of European tomato crops by Tuta absoluta: ecology, geographic expansion and prospects for biological control. J Pest Sci. 83:197–215.
- Desneux N, Luna MG, Guillemaud T, Urbaneja A. 2011. The invasive South American tomato pinworm, Tuta absoluta, continues to spread in Afro-Eurasia and beyond: the new threat to tomato world production. J Pest Sci. 84:403–408.
- Wolstenholme DR. 1992. Animal mitochondrial DNA: structure and evolution. Int Rev Cytol. 141:173–216.
- Yuan M, Yang H, Dai RH. 2019. Complete mitochondrial genome of Sitotroga cerealella (Insecta: Lepidoptera: Gelechiidae). Mitochondrial DNA B. 4:235–236.