Abstract
Litsea coreana var. lanuginose is an evergreen shrub or small tree of the genus Lauraceae. The complete chloroplast genome of L. coreana var. sinensis is 152,834 bp in length, containing an LSC region of 93,789 bp, an SSC region of 18,913 bp, and a pair of inverted repeats (IRA and IRB) of 20,066 bp each. The genome encodes of 127 genes, including 81 protein-coding genes, 8 ribosomal RNA genes, 36 transfer RNA genes, and 2 pseudogenes. Most of these genes occurred in a single copy, whereas 13 genes occurred in double copies, including all rRNA, 6 tRNA, and 3 protein-coding genes. The overall GC content is 39.1%. Phylogenetic analysis of cp genomes from 29 species of Lauraceae showed that cp genomes of L. coreana var. sinensis is most related to those of L. glutinosa, Laurus nobilis, and Lindera glauca.
Litsea coreana var. lanuginose is an evergreen shrub or small tree of the genus Lauraceae. Its bark is small scale-shape flaking, leaving deer skin mark on the trunk, so-called leopard-skin camphor tree in Guizhou province, China. The local people drink the tea made from its tender tips and leaves every day. As excessive leaf picking seriously affects the tree growth, the species is in an endangered state and urgently needs conservation.
The chloroplasts (cp) genome of higher plants is uniparental inheritance. Characterizing the chloroplast genome will facilitate the sustainable conservation of plant species. Therefore, in this study, we assembled and characterized the complete cp genome of L. coreana var. lanuginose.
The leaves of L. coreana var. sinensis were collected from Zhen'an County, Guizhou province of China (28°24′47.3″N, 107°27′40.6″E), and deposited in the Institute for Forest Resources & Environment of Guizhou, Guizhou University, China. The DNA was sequenced on the Illumina MiSeq platform. More than 4.5 Gb reads were obtained and trimmed with Trimmomatic v0.36 (Bolger et al. Citation2014), and then de novo assembly into contigs by CLC workbench. The longest contig was found to be a part of cp genome of L. coreana var. sinensis by BLAST search, and the highest homology cp genome comes from species L. glutinosa (Hinsinger and Strijk Citation2017); thus, L. glutinosa cp genome (GenBank: KU382356) was used as reference for construction of the whole cp genome of L. coreana var. sinensis by using MITObim v1.7 (Hahn et al. Citation2013). Then, the constructed cp genome was verified with the cp contigs from de novo assembly. The cp annotation was performed in GENEIOUS R10 (Kearse et al. Citation2012). The coding sequences, tRNAs and rRNAs were further confirmed and manually adjusted in some cases after BLAST searches. The annotated sequence was then deposited in GenBank under the accession number MK674781.
The complete cp genome of L. coreana var. sinensis is 152,834 bp in length, containing a LSC region of 93,789 bp, a SSC region of 18,913 bp, and a pair of inverted repeats (IRA and IRB) of 20,066 bp each. The genome encodes of 127 genes, including 81 protein-coding genes, 8 ribosomal RNA genes, 36 transfer RNA genes, and 2 pseudogenes. Most of these genes occurred in a single copy, whereas 13 genes occurred in double copies, including all rRNA (4.5S, 5S, 16S, and 23S rRNA), 6 tRNA (trnL-CAA, trnV-GAC, trnI-GAU, trnA-UGC, trnN-GUU, trnR-ACG), and 3 protein-coding genes (ndhB, rps7, rps12). The overall GC content is 39.1%.
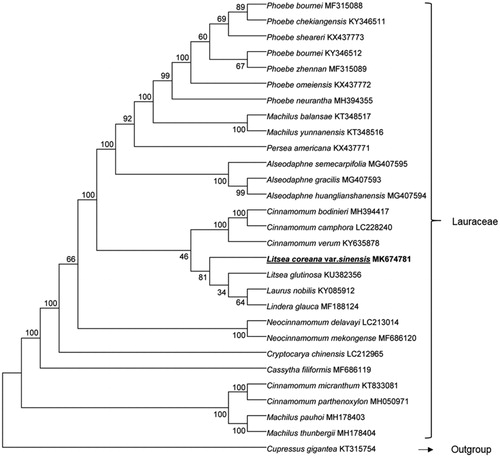
For the phylogenetic analysis, cp sequences from 28 species of Lauraceae were used. Sequences were aligned by multiple alignment in GENEIOUS R10, and a NJ tree was conducted using the MEGA 6.0 program (Tamura et al. Citation2013) with a bootstrap value of 1000. Phylogenetic tree shows that cp of L. coreana var. sinensis is most related to those of L. glutinosa, Laurus nobilis, and Lindera glauca, with a bootstrap support value of 81% (). The newly characterized chloroplast genome will provide basic data for the further conservation of L. coreana var. sinensis.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30:2114–2120.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads-a baiting and iterative mapping approach. Nucleic Acids Res. 41:e129.
- Hinsinger DD, Strijk JS. 2017. Toward phylogenomics of Lauraceae: the complete chloroplast genome sequence of Litsea glutinosa (Lauraceae), an invasive tree species on Indian and Pacific Ocean islands. Plant Gene. 9:71–79.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.