Abstract
The blackmouth croaker, Atrobucca nibe (Jordan & Thompson, 1911), is one of the most important economic fish in China. The mitogenome of A. nibe was determined in this study. The full circle length of the complete mitochondrial DNA was 16,505 bp. It contained 13 protein-coding genes, 22 tRNA genes, 2 rRNA genes, and a control region (D-loop). All 21 tRNA genes can fold into a typical cloverleaf structure except for tRNASer (AGY). The phylogenetic analysis revealed that A. nibe clustered into the three species of Sciaenidae (Pennahia microcephalus, Chrysochi aureus and Protonibea diacanthus).
Blackmouth croaker, Atrobucca nibe, is a fish belonging to the genus Sciaenidae of the family Perciformes (Caspers Citation1978). The family comprises ∼270 species in 70 genera in the world. Atrobucca nibe, as a commercially important group of fishes, occurs in West Pacific and inhabits along coastal waters from 45 to 200 m depth (Froese and Pauly Citation2019). In this study, the specimen of A. nibe sampled from East China Sea (122°20′31″, 32°0′11″) and was stored in the Key Laboratory of East China Sea Fishery Resources Exploitation, Ministry of Agriculture.
The complete mitochondrial genome sequence of A. nibe has been deposited in GenBank with an accession number (MK714015). The circle genome of A. nibe was 16,505 bp in length. The nucleotide composition of the heavy strand was 27.11% for A, 25.1% for T, 31.35% for C, and 16.44% for G. The A + T content (52.21%) was higher than the G + C content (47.79). It contained 22 transfer RNA genes (tRNA), 13 protein-coding genes, 2 rRNA genes (12S rRNA and 16S rRNA), and 1 control region. The length of 21 tRNAs ranged from 66 to 74 bp and their anti-codons were consistent with other fish of Sciaenidae. All of them could be folded into typical cloverleaf secondary structure except tRNASer(AGY). All genes were encoded on the heavy strand except ND6 and eight tRNAs. 13 protein-coding genes were initiated with the orthodox ATG. ND2, ATPase 6, ATPase 8, COIII, ND4L, ND5 and ND6 were terminated with TAA codon, ND1 and ND3 with TAG codon, COII with AGA codon, and the remaining genes with incomplete stop codon T––. The 12S and 16S rRNA genes were 953 bp and 1696 bp in length, respectively, and typically isolated by tRNAVal, located between tRNAPhe and tRNALeu. The D-loop is located between the tRNAPhe and tRNAThr genes with the size of 825 bp on the heavy strand.
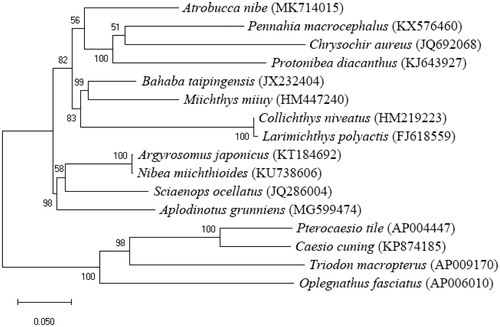
To investigate the phylogenetic relationship among drum fish, we downloaded the mitochondrial genome sequences of 15 currently available Eupercaria, including P. microcephalus (KX576460), C. aureus (JQ692068), P. diacanthus (KJ643927), Bahaba taipingensis (JX232404), Miichthys miiuy (HM447240), Collichthys niveatus (HM219223), Larimichthys polyactis (FJ618559), Argyrosomus japonicas (KT184692), Nibea miichthioides (KU738606), Sciaenops ocellatus (JQ286004), Aplodinotus grunniens (MG599474), Pterocaesio tile (AP004447), Caesio cuning (KP874185), Triodon macropterus (AP009170) and Oplegnathus fasciatus (AP006010). The concatenated nucleotide sequences of 13 protein-coding genes, 22 tRNAs, 2 rRNAs, and the D-loop of 16 species were aligned with the CLUSTAL W program(Larkin et al. Citation2007) and refined by eye. Phylogenetic trees were reconstructed with the concatenated nucleotide alignment using MEGA6 (Tamura et al. Citation2013) for maximum likelihood methods. Tree topology was evaluated by 1000 bootstrap replicates. The best-fit model to nucleotide substitution of these genomes was GTR + G + I. Phylogenetic analysis revealed that A. nibe clustered into the three species of Sciaenidae (P. microcephalus, C. aureus and P. diacanthus) ().
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Caspers H. (1978). Ethelwynn Trewavas: The Sciaenid fishes (Croakers or Drums) of the Indo-West-Pacific. — With 14 pl., 61 fig., 541 p. Transactions of the Zoological Society of London, Vol. 33, Part 4. London: Academic Press 1977, Vol 63.
- Froese R, Pauly D. (Editors). 2019. FishBase; [accessed February 2019]. www.fishbase.org.
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics. 23:2947–2948.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.