Abstract
Amomum villosum Lour. is a medicinal herb of high economic value, which is endemic to southern China. The size of A. villosum chloroplast (cp) genome was 163,733 bp long, which consisted of long (LSC; 88,798 bp) and short (SSC; 15,353 bp) single-copy regions, separated by a pair of inverted repeats (IRs; 29,791 bp for each unit). Totally, 126 genes are annotated in the cp genome, including 88 protein-coding genes (PCGs), 30 transfer RNA genes (tRNAs), and 8 ribosomal RNA genes (rRNAs). The overall GC content is 36.1%. The phylogenetic analysis based on 43 conserved protein-coding genes from 32 plant species recovered a consistent phylogeny on most of the nodes with previous results and indicated A. villosum as the sister species of the common ancestor of A. compactum and A. krervanh, and suggested that the cp genome data can effectively resolve the phylogenetic relationships. Our study provides a valuable resource for further study on its genetic diversification, evolution and germplasm conservation of this valued medicinal plant.
Amomum villosum Lour. is a medicinal herb of high economic value (Zhou Citation1993) in Zingiberaceae family due to its antioxidant (XuMei et al. Citation2018), analgesic, and anti-inflammatory effects (Wu et al. Citation2004). It has been used to treat gastrointestinal diseases in TCM (Traditional Chinese Medicine). Until recently, studies on A. villosum are mainly focused on the effective compounds or the genes encoding enzymes involved in producing medicinal natural products (Yang et al. Citation2012). Its complete chloroplast genome sequence is yet unavailable. Lack of genome information of A. villosum is a bottleneck for further genetic breeding and germplasm diversity and conservation studies. In this study, we sequenced and assembled the complete cp genome of A. villosum and analyzed the phylogeny.
The A. villosum plant used for genome sequencing was collected from cultivated fields in Yangchun, Guangdong province, China. The specimen and DNA of A. villosum is stored in University of Macau (Specimen accession number HYW170002). To extract DNA from fresh and healthy leaves, we used DNeasy Plant Mini Kit (QIAGEN). The total DNA concentration was estimated by measuring A260/A280 with Qubit Fluorometric Quantification (Thermo Fisher). Then, we used BGISEQ500 sequencing technology according to the manufacturers’ kits. The raw reads were filtered by SOAPnuke (SOAPnuke, RRID:SCR_015025) (Chen et al. Citation2018). Finally, the clean reads were ∼30× coverage of the whole cp genome.
The complete cp genome of A. villosum is 163,733 bp in length (GenBank accession number: MK389642) with a typical circular structure of cellular organelle genome, including a pair of IR regions (each of 29,791 bp) separated by LSC (88,798 bp) and SSC (15,353 bp) regions. Overall, the CG content of A. villosum cp genome is 36.1%, with the highest GC content in the IRs regions (41.2%) than in LSC and SSC (33.7% and 30%, respectively). There are totally 126 genes in the genome, including 88 protein-coding genes, 30 tRNA genes, and 8 rRNA genes.
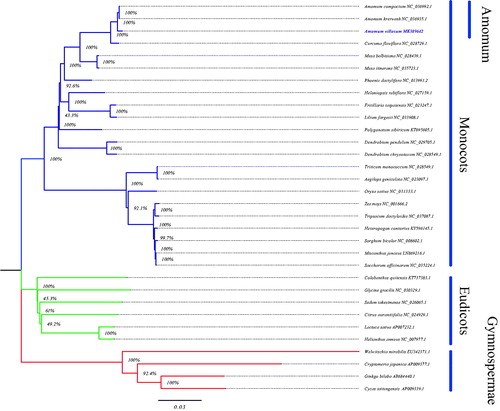
Phylogenetic analysis was conducted based on sequences from 32 taxa, including 22 monocots, 6 eudicots, and 4 gymnospermae as outgroups. First, the sequences of the 43 conserved protein-coding genes were extracted from these 32 species cp genomes (downloaded from NCBI) by Sequence Manipulation Suite (http://www.bioinformatics.org/sms2/genbank_feat.html) and Perl scripts. Second, these conserved genes were aligned using MUSCLE3.8.31 (Edgar Citation2004) with default settings. Finally, a neighbor-joining (NJ) tree was constructed in MEGA v7 (Kumar et al. Citation2016) with 1000 bootstrap replicates. Bootstrap analysis shows that 25 of the 29 nodes are supported by values over 90%, and 21 nodes support by 100% bootstrap value (). The nodes between the three species of Amomum genus are all supported by bootstrap values of 100% in NJ tree. These phylogenetic results strongly support the position of A. villosum as a sister species of A. compactum and A. krervanh, which are also herbal medicine in China. Generally speaking, the phylogenetic results infering from the cp genome data are highly supportive and are similar to the results obtained from nuclear genes data, showing that the cp genome data can effectively resolve the phylogenetic relationships.
Disclosure statement
The authors declare no conflicts of interest and are responsible for the content.
Additional information
Funding
References
- Chen Y, Chen Y, Shi C, Huang Z, Zhang Y, Li S, Li Y, Ye J, Yu C, Li Z, et al. 2018. SOAPnuke: a MapReduce acceleration-supported software for integrated quality control and preprocessing of high-throughput sequencing data. Gigascience. 7:1–6.
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- Wu X, Li X, Xiao F, Zhang Z, Xu Z, Wang H. 2004. [Studies on the analgesic and anti-inflammatory effect of bornyl acetate in volatile oil from Amomum villosum]. Zhong Yao Cai. 27:438–439.
- XuMei Z, MingZhong C, JiaMing P. 2018. Antioxidant and antibacterial activities of the crude extract of total flavonoids from Amomum villosum Lour. J Food Safety Qual. 9:3335–3339.
- Yang J, Adhikari MN, Liu H, Xu H, He G, Zhan R, Wei J, Chen W. 2012. Characterization and functional analysis of the genes encoding 1-deoxy-d-xylulose-5-phosphate reductoisomerase and 1-deoxy-d-xylulose-5-phosphate synthase, the two enzymes in the MEP pathway, from Amomum villosum Lour. Mol Biol Rep. 39:8287–8296.
- Zhou S. 1993. Cultivation of Amomum villosum in tropical forests. Forest Ecol Manage. 60:157–162.