Abstract
In this work, we reported the complete mitochondrial genome of Corydoras duplicareus. The total length of the mitochondrial genome is 16,667 bp, with the base composition of 32.90% A, 26.55% T, 25.94% C, and 14.61% G. It contains two ribosomal RNA genes, 13 protein-coding genes, 22 transfer RNA genes, and a major non-coding control region (D-loop region). The arrangement of these genes is the same as that found in the Siluriformes. All the protein initiation codons are ATG, except for cox1 that begins with GTG. The cytochrome c oxidase subunit I (COX1) amino acid sequence of C. duplicareus and other 27 species from 8 genera were used for phylogenetic analysis by maximum likelihood method. The topology demonstrated that all species are divided into two groups (Callichthyidae and Cyprinidae), and the C. duplicareus was clustered with C.nattereri.
Corydoras duplicareus, which belongs to the genus Corydoras within the catfish family Callichthyidae, was distributed in South America. Now many species in the Callichthyidae family are introduced to China as the kind of small ornamental fish. But, the naming and classification of these ornamental catfish is not accurate in China. As we know, the mitochondrial genome can be applied to research areas such as species identification, taxonomy, and phylogeny of fish (Akihito et al. Citation2000; Keith et al. Citation2011).
In this study, we determined the complete mitochondrial DNA sequence of C. duplicareus to provide the molecular biology classification basis. The samples which preserved in 99% ethanol in Museum of Hunan Agricultural University, were collected from Yangfan ornamental fish market in changsha, Hunan province (113°04E, 28°20N) in November 2015. The genomic DNA was isolated from fin tissue using the EasyPure Genomic DNA Kit (Transgen Biotech, Beijing, China) according to the manufacturer’s protocol. 22 primer pairs were used to amplify the complete mitochondrial genome by the PCR. The PCR products were detected by agarose gel electrophoresis and sequenced by the company Shanghai BioSune. DNA sequences were analyzed as that presented in the previous studies (Liu et al. Citation2013) and annotated by software Geneious R11. The complete mitochondrial genome sequence of C. duplicareus is 16,667 bp in length (GenBank accession number: MK675098), containing 2 ribosomal RNA genes, 13 protein-coding genes, 22 transfer RNA genes, and a major non-coding control region 1053 bp in length (D-loop region). The arrangement of these genes is the same as that found in the Siluriformes. All the protein initiation codons are ATG, except for cox1 that begins with GTG.
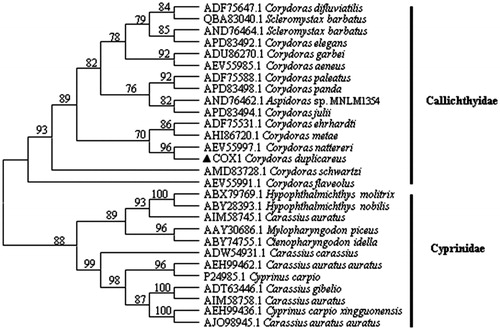
The complete mitochondrial genome sequence of the family Callichthyidae is really limited, and the only two records in GenBank were C. rabauti and C. nattereri. The cytochrome c oxidase subunit I (COX1) amino acid sequence of C. duplicareus and other 27 sequence from 8 genera were used for phylogenetic analysis by maximum likelihood method (bootstrap value was 1000) in this study (Botero-Castro et al. Citation2016). The phylogenetic tree was reconstructed with MEGA6. 16 sequence from 3 genera (Aspidoras, Corydoras and Scleromystax) belong to Callichthyidae, and 12 sequence from 5 genera (Hypophthalmichthys, Mylopharyngodon, Ctenopharyngodon, Carassius and Cyprinus) belong to Cyprinidae. The topology the phylogenetic tree in this study () demonstrated that all species are divided into two groups (Callichthyidae and Cyprinidae), and the C. duplicareus was clustered with C. nattereri. The results of this study confirmed the classified status of C. duplicareus from Hunan China and provided the molecular biology basis for classification.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Akihito, Iwata A, Kobayashi T, Ikeo K, Imanishi T, Ono H, Umehara Y, Hamamatsu C, Sugiyama K, Ikeda Y, Sakamoto K, et al. 2000. Evolutionary aspects of gobioid fishes based upon a phylogenetic analysis of mitochondrial cytochrome b genes. Gene. 259:5–15.
- Botero-Castro F, Delsuc F, Douzery E. 2016. Thrice better than once: quality control guidelines to validate new mitogenomes. Mitochondrial DNA Part A. 27:449–454.
- Keith P, Lord C, Lorion J, Watanabe S, Tsukamoto K, Couloux A, Dettai A. 2011. Phylogeny and biogeography of Sicydiinae (Teleostei: Gobiidae) inferred from mitochondrial and nuclear genes. Mar Biol. 158:311–326.
- Liu Q-L, Xu B-H, Xiao T-Y, Su J-M, Yao Y-B, Liu Y-J. 2013. Complete mitochondrial genome of the Xiangjiang barbel chub Squaliobarbus curriculus: comparative analysis of the genetic variation associated with geographical population. Mitochondrial DNA. 24:654–656.