Abstract
Magnolia omeiensis (W. C. Cheng) Dandy, belonging to Magnolia sect. Gynopodium Dandy (Magnoliaceae), is a critically endangered species. In this study, we assembled the complete plastid genome sequence of M. omeiensis. The plastid genome was 160,091 bp in length, consisting of a large single-copy (LSC) region of 88,160 bp, a small single-copy (SSC) region of 18,763 bp, and two inverted repeat (IR) regions of 26,584 bp. It contained 112 unique genes, including 80 protein-coding genes, 28 tRNA genes, and 4 rRNA genes. Overall GC content was 39.3%. Phylogenetic analysis showed that M. omeiensis was closely related to Magnolia Yunnanensis, which was identified in the same section Gynopodium.
Magnolia omeiensis (W. C. Cheng) Dandy, belonging to Magnolia sect. Gynopodium Dandy (Magnoliaceae) (Figlar and Nooteboom Citation2004), is endemic to China, and has an extremely restricted distribution range in Emei Mountain (Xiao and Li Citation2017). Due to habitat loss, M. omeiensis was categorized as Critically Endangered plant to China, according to the latest version of IUCN Red List of threatened species (China Expert Workshop Citation2014). Conservation studies on M. omeiensis have mainly focused on phytocoenological characteristics and breeding (Zhang et al. Citation1993; Wade et al. Citation2016; Yu et al. Citation2018). In addition, genetic structure and phylogeographical patterns of sect. Gynopodium from subtropical China were detected by using chloroplast psbA-trnH sequences (Xiao and Li Citation2017). Understanding the plastid genome of M. omeiensis is also crucial for its conservation concerns, but so far there is no plastid genome resource for this species. Therefore, we assembled the whole plastid genome of M. omeiensis in this study.
Voucher specimen (SCBG00000853, IBSC) and leaves of M. omeiensis were sampled from South China Botanical Garden (N23°11′, E113°22′). Total genomic DNA was extracted from leaves dried by silica gel, then sequenced on an Illumina Hiseq X Ten platform. c.2 Gb of paired-end (PE = 150 bp) sequence reads were obtained and was used to assemble the plastid genome by NOVOPlasty 2.7.2 (Dierckxsens et al. Citation2017). The complete plastid genome sequence of Magnolia yunnanensis (GenBank Accession Number KF753638) was set as a seed sequence in the seed-and-extend algorithm implemented in NOVOPlasty. The genes in the plastid genome were annotated using the online tool GeSeq (Tillich et al. Citation2017), with manual adjustment using Geneious 11.0.2 (Kearse et al. Citation2012). ARAGORN (Laslett and Canback Citation2004) was used to detect transfer RNA (tRNA). The annotated plastid genome of M. omeiensis has been deposited in GenBank (Accession Number: MK713973).
The complete plastid genome of M. omeiensis was 160,091 bp in length. It typically consisted of a large single-copy (LSC) region of 88,160 bp, a small single-copy (SSC) region of 18,763 bp, and a pair of inverted repeat (IRa and IRb) of 26,584 bp. Overall GC content was 39.3%. The plastid genome contains 112 unique genes, including 80 protein-coding genes, 28 tRNA genes, and 4 ribosomal RNA (rRNA) genes.
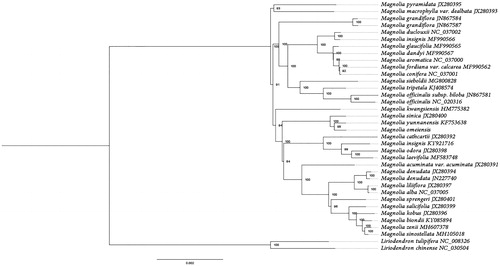
Phylogenomic analysis was performed using the complete plastid genomes from 34 samples in the subfamily Magnolioideae (including M. omeiensis in this study). Liriodendron tulipifera and Liriodendron chinense from subfamily Liriodendroidae were used as outgroups. Whole plastid genome sequences from all the above-menioned species were aligned using MAFFT (Katoh and Standley Citation2013). Maximum likelihood phylogeny was constructed using RAxML (Stamatakis Citation2014) with GTR + G model. The bootstrap support values were based on 1000 replicates. The phylogeny tree showed that M. omeiensis was closely releated to Magnolia Yunnanensis (), which was identified in the same section Gynopodium (Figlar and Nooteboom Citation2004). The plastid genome of M. omeiensis is of significance for its conservation and evolutionary studies.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- China Expert Workshop. 2014. Magnolia omeiensis. The IUCN Red List of Threatened Species 2014: e.T34961A2857537.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:e18.
- Figlar RB, Nooteboom HP. 2004. Notes on Magnoliaceae IV. BLUMEA. 49:87–100.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Laslett D, Canback B. 2004. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 32:11–16.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics (Oxford, England). 30:1312–1313.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45:W6–W11.
- Wade EM, Nadarajan J, Yang X, Ballesteros D, Sun W, Pritchard HW. 2016. Plant species with extremely small populations (PSESP) in China: a seed and spore biology perspective. Plant Divers. 38:209–220.
- Xiao LQ, Li QQ. 2017. Phylogeography and allopolyploidization of Magnolia sect. Gynopodium (Magnoliaceae) in subtropical China. Plant Systemat Evol. 303:957–967.
- Yu DP, Li CH, Wen XY, Li XJ, Peng QX, Xie KP. 2018. Flowering biology and breeding system of Parakmeria omeiensis. Guihaia. DOI:10.11931/guihaia.gxzw201804021
- Zhang P, Liu RY, Liang KH. 1993. A preliminary study on phytocoenological characteristics of Parakmeria omeiensis community. Guihaia. 13:61–69.