Abstract
Known as a high quality forage grass, Achnatherum splendens distributes in the northwest of China, northeast, Inner Mongolia, shanxi and hebei provinces. In this study, we assembled the complete chloroplast (cp) genome of A. splendens using data from high-throughput Illumina sequencing. The A. splendens cp genome is 136,876 bp in size and includes two inverted repeat regions of 21,639 bp each, which is separated by a large single copy region of 80,958 bp and a small single copy region of 12,640 bp. A total of 130 genes were predicted, including 38 tRNA, 8 rRNA, and 84 protein-coding genes. In addition, 8 PCG genes possess a single intron, 74 PCG genes no intron, 2 other genes harbor two introns. 6 tRNA genes harbor a single intron. Phylogenetic analysis placed A. splendens within the Stipeae.
The Achnatherum splendens (Trin.) Nevski, is a high quality forage grass with thick leathery outer sheathed fibrous roots (Chen et al. Citation1987). As the best-known species in the family pooideae, A. splendens distributes in the northwest of China, northeast, Inner Mongolia, shanxi and hebei provinces. It can grow on slightly alkaline sandy beaches and sandy slopes and improve alkaline land, protect canal and conserve water and soil. At present, there are uncertainties in the classification of pooideae, compromising the understanding of evolution within the family (Liu et al. Citation2016; Tsuruta et al. Citation2017) and hampering conservation efforts of its member species. Currently, there is little complete chloroplast (cp) genome sequence of poaceae species was reported (Saarela et al. Citation2015). We aimed to assemble and characterize A. splendens’s cp genome to provide a better understanding on the evolution and genetics of A. splendens and other species in the same family.
Total genomic DNA was extracted from fresh leaves collected from Yinchuan Belly Desert (38°28′N, 106°16′E; Ningxia, NW China). A voucher specimen (P02B) is deposited at Breeding Base for State Key Laboratory of Land Degradation and Ecological Restoration of North-western China in Ningxia University. A genomic library with an insert size of _400 bp was prepared using a TruSeq DNA Sample Prep Kit (Illumina, Beijing) and sequenced on an Illumina HiSeq X Ten platform. Approximately 8 Gb of raw data were generated through pair-end 150 bp sequencing. After removal of adapter sequences, raw reads were fed into the NOVOPlasty (Dierckxsens et al. Citation2017) for assembly with the rbcL gene of Stipa hymenoides (GenBank accession KM974729) as the seed sequence. The assembled cp genome was annotated using the online annotation tool DOGMA (Wyman et al. Citation2004) and further corrected manually. The complete cp genome of A. splendens (GenBank accession MK704435) was 136,876 bp in length, consisting of a pair of inverted repeat regions of 21,639 bp each, a large single copy region of 80,958bp, and a small single copy region of 12,640 bp. A total of 130 genes were annotated, including 38 tRNA, 8 rRNA, and 84 protein-coding genes. The overall GC content of the cp genome was 38.9%. In addition, 8 PCG genes (rps16, atpF, petB, petD, rpl16, ndhA, ndhB and rpl2) possess a single intron, 74 PCG genes no intron, 2 other genes (rps12 and ycf3) harbor two introns. 6 tRNA genes (trnA-UGC, trnI-GAU, trnK-UUU, trnG-UUC, trnL-UAA and trnV-UAC) harbor a single intron.
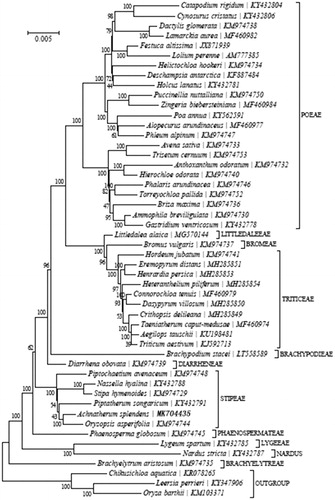
A phylogenetic analysis was carried out with A.splendens and 46 other complete cp genomes of species from the Pooideae, comprising of 11 Tribus (Poeae, Triticeae, Littledaieeae, Bromeae, Brachypodieae, Diarrheneae, Stipeae, Phaenospermateae, Lygeeae, Nardus and Brachyelytreae) and three other species (Chikusichloa aquatic, Leersia perrieri and Oryza barthii) was included as outgroup. The maximum likelihood tree constructed with RAxML (Stamatakis Citation2014), implemented in Geneious ver. 10.1 (http://www.geneious.com, Kearse et al. Citation2012), placed A.splendens within the Stipeae ().
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Chen XQ, Cui HB, Dai LK. 1987. Flora of China. Vol. 9. Beijing: Science Press.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:e18.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Liu F, Tembrock LR, Sun C, Han G, Guo C, Wu Z. 2016. The complete plastid genome of the wild rice species Oryza brachyantha (Poaceae). Mitochondrial DNA Part B. 1:218–219.
- Saarela JM, Wysocki WP, Barrett CF, Soreng RJ, Davis JI, Clark LG, Kelchner SA, Pires JC, Edger PP, Mayfield DR, et al. 2015. Plastid phylogenomics of the cool-season grass subfamily: clarification of relationships among early-diverging tribes. AoB Plants. 7:plv046.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Tsuruta S-i, Ebina M, Kobayashi M, Takahashi W. 2017. Complete chloroplast genomes of Erianthus arundinaceus and Miscanthus sinensis: comparative genomics and evolution of the Saccharum complex. PLoS One. 12:e0169992.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.