Abstract
Cyperus difformis is a pioneering plant of aquatic and moist habitats, which is widely distributed in southern Europe and plays an important role in sand fixation, soil improvement, and water purification. Here, the complete chloroplast genome of the C. difformis has been reconstructed from the whole-genome Illumina sequencing data. The circular genome is 167,974 bp in size, and comprises a pair of inverted repeat (IR) regions of 38,040 bp each, a large single-copy (LSC) region of 83,484 bp and a small single-copy (SSC) region of 8,410 bp. The total GC content is 36.1%, while the corresponding values of the LSC, SSC and IR region are 34.6%, 27.7%, and 38.6%, respectively. The chloroplast genome contains 137 genes, including 91 protein-coding genes, 8 ribosomal RNA genes, and 38 transfer RNA genes. The maximum-likelihood phylogenetic analysis showed a strong sister relationship with Carex plants in Cyperaceae. Our findings provide a foundation for further investigation of chloroplast genome evolution in C. difformis and other pioneering plants.
Cyperus difformis is aquatic and moist habitats that are native to southern Europe. It is rich in essential oils and has high photosynthetic efficiency, which could help in becoming a pioneering plant in the poor environment (Akter et al. Citation2018; Erdem et al. Citation2018). Also, C. difformis play an important role in phytoremediation of heavy metal pollution in water and soils (Vara Prasad and de Oliveira Freitas Citation2003; Anoliefo et al. Citation2008; Chang et al. Citation2009). So far, little genetic information on C. difformis is reported. Here, we assembled and annotated the complete chloroplast genome of C. difformis using high-throughput sequencing technology, which will be helpful for further studies on the function of chloroplast genes and phylogenetic evolution
The genomic DNA was extracted from a single individual C. difformis growing in Zhalong Wetlands, Qiqihar of Heilongjiang province, China (N47°12′ E124°14′). DNA samples of C. difformis were stored in Heilongjiang Province Key Laboratory of Geographical Environment Monitoring and Spatial Information Service in Cold Regions, Harbin Normal University, Harbin, China. High-throughput DNA sequencing was conducted on the Illumina HiSeq 2500 Sequencing System (Illumina, CA, USA) by Shanghai Genesky Biotechnologies Inc (Shanghai, China). Total 35.18 M raw reads were retrieved and trimmed by CLC Genomics Workbench v8.0 (CLC Bio, Aarhus, Denmark), and 23.05 M reads were aligned and ordered to the reference cp genome of Hypolytrum nemorum (GenBank: NC_036036.1) by NOVOPlasty (Dierckxsens et al. Citation2016). The chloroplast genome was annotated in GENEIOUS R11 (Biomatters Ltd., Auckland, New Zealand).
The chloroplast genome of C. difformis is a circular DNA molecule with 167,974 bp in size (MK423991). It comprises a pair of inverted repeat (IR) regions of 38,040 bp each, separated by a large single-copy (LSC) region of 83,484 bp and a small single-copy (SSC) region of 8,410 bp. The total GC content of the chloroplast genome is 36.1%. The chloroplast genome harbors 137 functional genes, including 91 protein-coding genes (PCGs), 38 tRNA genes, and 8 rRNA genes. Among them, 53 are involved in photosynthesis, and 70 genes are involved in self-replication. Moreover, among all the protein-coding genes, 12 genes contain one intron, while atpA, chlp, and ycf3 harbor two introns.
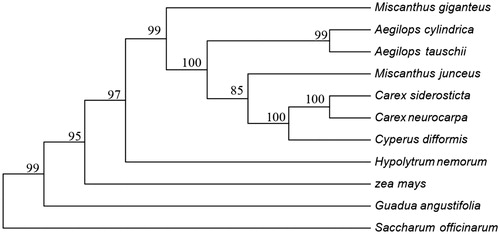
To determine the phylogenetic position of C. difformis, 53 photosynthesis related PCGs sequences among 11 chloroplast genomes were aligned by MAFFT (Katoh et al. Citation2002) and then were connected as gene strings. The maximum-likelihood phylogenetic tree of C. difformis was generated using those gene strings sequence by MEGA 6.0 (Tamura et al. Citation2013) using 1000 bootstrap replicates. The phylogenetic analysis () showed the position of C. difformis was situated as the sister of C. siderosticta and C. neurocarpa in Cyperaceae. Our findings provide valuable information on the genetic diversity research of C. difformis and enrich the resources of chloroplast genomes in pioneering plants.
Figure 1. Phylogenetic of 10 species based on the maximum-likelihood analysis of the whole chloroplast genome sequences using 500 bootstrap replicates. The analyzed species and corresponding Genbank accession numbers are as follows: Aegilops cylindrica (NC_023096), Aegilops tauschii (NC_022133), Carex siderostict (NC_027250), Carex neurocarpa (KU238086), Guadua angustifolia (NC_029749), Hypolytrum nemorum (NC_036036), Miscanthus x giganteus (NC_035753), Miscanthus junceus (NC_035751), Saccharum officinarum (LN849913), Zea mays (NC_001666).
Disclosure statement
No potential conflict of interest was reported by the author.
Additional information
Funding
References
- Akter F, Begum M, Salam A. 2018. In situ and ex situ floristic diversity of weed seedbank in rice at farmers’ fields. J Res Weed Sci. 2:75–89.
- Anoliefo GO, Ikhajiagbe B, Okonokhua BO, Edegbai BE, Obasuyi OC. 2008. Metal tolerant species distribution and richness in and around the metal based industries: possible candidates for phytoremediation. Afr J Environ Sci Technol. 11:360–370.
- Chang HQ, Kou TJ, Qiao XH, He JY. 2009. Removal efficiency of nutrients from wastewater by several plants. Bull Soil Water Conserv. 5:29–34.
- Dierckxsens N, Mardulyn P, Smits G. 2016. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 4:e18–e18.
- Erdem B, Bagci E, Dogan G, Aktoklu E, Dayangac A. 2018. Chemical composition and antimicrobial activities of essential oil and ethanol extract of Cyperus fuscus L burs from Turkey. Trop J Pharm Res. 17:1637–1643.
- Katoh K, Misawa K, Kuma K, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30:3059–3066.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- Vara Prasad MN, de Oliveira Freitas HM. 2003. Metal hyperaccumulation in plants: biodiversity prospecting for phytoremediation technology. Electron J Biotechnol. 3:285–321.
