Abstract
Cirsium japonicum (C. japonicum) is an important oriental herb belonging to the family Asteraceae. The C. japonicum complete chloroplast genome is composed of 152,606 bp, which form a large single-copy region (LSC, 83,492 bp), a small single-copy region (SSC, 18,772 bp), and 2 inverted repeats (IRs, 25,196 bp). There are 114 genes annotated, including 80 protein-coding genes, 4 ribosomal RNA genes, and 30 transfer RNA genes. Our phylogenetic analysis suggests that C. japonicum is closely related to the Cirsium genus of the Asteraceae family, and that C. japonicum (subfamily: Carduoideae) is separate from the Asteroideae subfamily.
Cirsium japonicum is a perennial, wild plant that belongs to the family Asteraceae. This plant has long been utilized in Eastern medicine as an anti-inflammatory, antioxidant, anti-tumor, and hepatitis agent in China, Japan, and Korea (Liu et al. Citation2006; Shin et al. Citation2017). We extracted genomic DNA from young C. japonicum leaves and whole body specimens that were registered to the Jeollabukdo Medicinal Resource Research Institute (geographic coordinate: N 35°46′10″, E 127°22′40″) under the voucher number JA20180995. A total of 5.12 Gbp of data were obtained using the Illumina MiSeq platform (San Diego, CA, USA), and trimmed reads were assembled using the CLC Assembly Cell software package (CLC Inc., Aarhus, Denmark). The selected contigs were merged and gap-filled by a series of read maps (Kim et al. Citation2015). The assembled chloroplast genome was annotated using the GeSeq program (Tillich et al. Citation2017), and the complete chloroplast genome was deposited into GenBank under accession number MH778960.
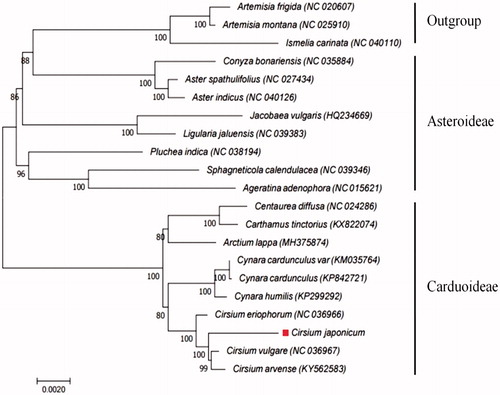
The C. japonicum chloroplast genome is 152,606 bp in length and has a circular structure with a large single-copy (LSC) region of 83,492 bp, a small single-copy (SSC) region of 18,772 bp, and a pair of 25,196 bp inverted repeats (IRa/IRb). Genome annotation revealed 114 functional genes, including 80 protein-coding genes, 4 ribosomal RNAs, and 30 transfer RNAs. Most of these are single-copy genes; however, 22 genes (9 protein-coding genes, 5 ribosomal RNA genes, and 8 transfer RNA genes) are located within the inverted repeat regions. A phylogenetic analysis was performed comparing the complete chloroplast genome against 20 Asteraceae family species, including 9 from the Carduoideae subfamily, 8 from the Asteroideae subfamily, and 3 outgroups. Phylogenetic relationships were revealed in comparisons of 59 common protein-coding gene sequences. The phylogeny indicated that C. japonicum species is more closely related to the Cirsium genus than the Cynara genus and that C. japonicum (subfamily: Carduoideae) is separate from the Asteroideae subfamily (). Results from this study can be used to resolve problems related to the phylogeny of genus Cirsium.
Figure 1. Phylogeny of C. japonicum and 20 related species based on complete chloroplast genome sequences. The phylogenetic tree was constructed using the maximum likelihood (ML) method with 1000 bootstrap replicates based on the complete chloroplast genomes of 21 species from the Asteraceae family, including 9 from the Carduoideae subfamily, 8 from the Asteroideae subfamily, and 3 outgroups.
Geolocation
The genomic DNA sample used for sequencing was obtained from the Jeollabukdo Medicinal Resource Research Institute (geographic coordinate: N 35°46′10″, E 127°22′40″).
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Kim K, Lee SC, Lee J, Yu Y, Yang K, Choi BS, Koh HJ, Waminal NE, Choi HI, Kim NH, et al. 2015. Complete chloroplast and ribosomal sequences for 30 accessions elucidate evolution of Oryza AA genome species. Sci Rep. 5:15655.
- Liu S, Luo X, Li D, Zhang J, Qiu D, Liu W, She L, Yang Z. 2006. Tumor inhibition and improved immunity in mice treated with flavone from Cirsium japonicum DC. Int Immunopharmacol. 6:1387–1393.
- Shin MS, Park JY, Lee J, Yoo HH, Hahm DH, Lee SC, Lee S, Hwang GS, Jung K, Kang KS. 2017. Anti-inflammatory effects and corresponding mechanisms of cirsimaritin extracted from Cirsium japonicum var. maackii Maxim. Brain Res Bull. 27:3076–3080.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES1, Fischer A, Bock R, Greiner S. 2017. GeSeq–versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45:W6–W11.
