Abstract
In this study, the complete mitochondrial genome of Grey-streaked Flycatcher (Muscicapa griseisticta) has been determined by the polymerase chain reaction method for the first time using the muscle tissue. The overall base composition of grey-streaked flycatcher mitogenome is 29.3% for A, 31.9% for C, 14.9% for G, and 23.9% for T. The percentage of G + C content is 46.8%. The total length of the sequence is 17,897 bp, which contains 13 protein-coding genes, 22 transfer RNA genes, 2 ribosomal RNA genes, 2 control regions. Phylogenetic analysis on the basis of 12 protein-coding genes and 2 rRNA genes of 16 passeriformes species’ mitochondrial genomes using maximum likelihood (ML) demonstrated that Oriental Magpie Robin (Copsychus saularis) is the most closest to Grey-streaked Flycatcher.
The Grey-streaked Flycatcher is a kind of passerine in the family Muscicapidae, which is mainly distributed in Northeast Asia, Indonesia, Malaysia, and Philippines. Species in Muscicapidae are finches and the classification of Muscicapidae is controversial. In this study, the complete mitochondrial genome sequence of Muscicapa griseisticta was sequenced by Chain Termination Method and reported for the first time using muscle tissue. The sample of M. griseisticta was collected from Yancheng City, Jiangsu Province, China. After sampling, the specimens were stored in animal specimens museum of Nanjing Forestry University. All sampling procedures and experiments were approved.
The complete mitogenome of M. griseisticta (Genbank accession number MK390479) was sequenced to be 17,897 bp. Sequence analysis showed that the genome structure is consistent with other Passeriformes species, which contains 13 protein-coding genes (PCGs), 22 transfer RNA (tRNA) genes, 2 ribosomal RNA (rRNA) genes, and 2 control regions. The structure of the mitogenome is a typical vertebrate mitochondrial gene arrangement (Boore Citation1999; Sun et al. Citation2016; Zhao et al. Citation2016).
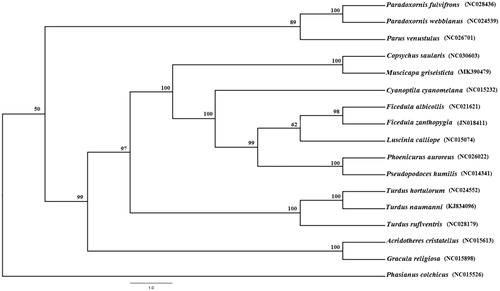
We used the maximum-likelihood (ML) method based on the 12 protein-coding genes (except ND6 gene) and 2 rRNA genes to analyze the phylogenetic relationships of M. griseisticta and 15 other Passeriformes species (). MrModeltest 3.7 was used for screening model, and GTR + I+G was chosen as the optimal evolutionary model according to the AIC test criterion (Tracey et al. Citation2007). Maximum parsimony and bootstrap consensus phylogenetic trees were computed with 1000 bootstrap replicate using PAUP* 4.0 (Florida State University, USA) (Felsenstein Citation1981).
In this tree, M. griseisticta is clustered with Copsychus saularis, which indicates that they have a much closer relationship (Zuccon et al. Citation2012; Questiau et al. Citation1999). The phylogenetic tree reveals that the data of our new determined mitogenome explain some related evolution issues. It appears that Muscicapidae and Turdidae are sister groups, the phylogenetic analysis result was consistent with the previous research (Zhang et al. Citation2016).
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Boore JL. 1999. Animal mitochondrial genomes. Nucleic Acids Res. 27:1767–1780.
- Felsenstein J. 1981. Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol. 17:368–376.
- Questiau S, Gielly L, Clouet M, Taberlet P. 1999. Phylogeographical evidence of gene flow among common crossbill (Loxia curvirostra, Aves, Fringillidae) populations at the continental level. Heredity. 83:196–205.
- Sun GL, Xia T, Yang XF, Zhao C, Liu GS, Sha WL, Zhang HH. 2016. The complete mitochondrial genome sequence of Eophona migratoria (Passeriformes Fringillidae). Mitochondr DNA B. 1:753–754.
- Tracey SR, Smolenski A, Lyle JM. 2007. Genetic structuring of Latris lineata at localized and transoceanic scales. Marine Biol. 152:119–128.
- Zhang H, Cheng YY, Zhou LZ, Dong YQ. 2016. Complete mitochondrial genome of white-throated rock-thrush Monticola gularis (Passeriformes: muscicapidae). Mitochondr DNA B. 1:684–685.
- Zhao C, Zhang HH, Liu GS, Yang XF, Zhang J. 2016. The complete mitochondrial genome of the Tibetan fox (Vulpes ferrilata) and implications for the phylogeny of Canidae. C R Biol. 339:68–77.
- Zuccon D, Prŷs-Jones R, Rasmussen PC, Ericson P. 2012. The phylogenetic relationships and generic limits of finches (Fringillidae). Mol Phylogenet Evol. 62:581–596.