Abstract
The complete chloroplast genome sequence of Lilium lankongense Franchet is presented here. It is 152,611 bp in length and divides into four distinct regions: small single copy region of 17,506 bp, large single copy region of 81,995 bp, and a pair of inverted repeat regions (26,555 bp). The L. lankongense chloroplast genome annotation predicted a total of 131 genes, which contains 83 protein-coding genes, 38 transfer RNA genes, and 8 ribosomal RNA genes. In the maximum likelihood tree, all kinds of Lilium were clustered into two monophyletic groups.
Keywords:
Lilium lankongense Franchet belonging to the genus Lilium (Liliaceae), is an herbaceous plant species naturally distributed in China (southeast Xizang, northwest Yunnan). It grows in alpine grasslands at an elevation from 1800 to 3200 m (Liang and Tamura Citation2000). Lilium lankongense possesses pink tepals with deep red spots, revolute margin, which has high ornamental value. It has attracted the attention of plant breeders and become good parents for interspecific hybridization (North and Wills Citation1969). Although several phylogenetic studies used sequences derived from nuclear and chloroplast of L. Lankongens (Nishikawa et al. Citation1999; Gao et al. Citation2012, Citation2013, Citation2015), available genetic resource currently for this plant is still scarce. Therefore, developing other genetic resources is necessary to protect and use the germplasm of this species. Here, we generated the complete chloroplast (cp) genome of L. lankongense for further research. The sequence was submitted to GenBank with the accession number MK757466.
Total genomic DNA was extracted from the mature and healthy leaves of a single individual of L. lankongense sampled from Lijiang (30°01' 56.6" N 101°50' 48.6" E), Yunnan province, China. Voucher specimens were deposited in SZ (Sichuan University Herbarium, herbarium number: KS2018071601). The obtained DNA was constructed into average 350 bp paired-end (PE) library by Illumina Hiseq platform (Illumina, San Diego, CA, USA) and sequenced using Illumina genome analyzer (Hiseq PE150Illumina, San Diego, CA,USA). The complete cp genomes were assembled with NOVOPlasty (Dierckxsens et al. Citation2016). Sequence annotation was accomplished using Geneious (Kearse et al. Citation2012) and compared with the chloroplast sequence of Lilium duchartrei as a reference.
The complete chloroplast genome of L. lankongense is a typical quadripartite structure of 152,611 bp in total length with 37.00% GC contents, including a pair of inverted repeats (IRs 26,555bp) separated by the large single copy region (LSC 81,995 bp) and small single copy region (SSC 17,506 bp). There are a total of 131 genes in the chloroplast genome, comprising 83 protein-coding genes, 38 transfer RNA genes, and 8 ribosomal RNA genes.
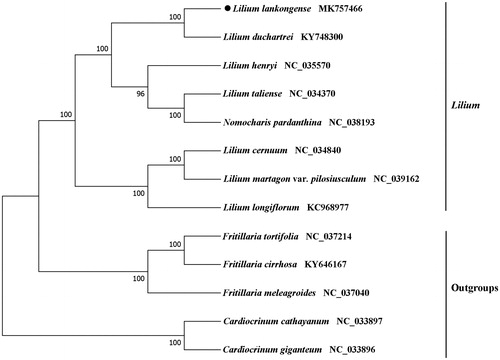
Phylogenetic analysis was constructed based on the complete chloroplast genome sequence obtained from L. lankongense with those of 7 reported species in the Lilium genus and five out-groups using the maximum likelihood (ML) analysis by RAxML 8.0 software (Stamatakis Citation2014). The branch support values were calculated from 1000 bootstrap replicates. In the phylogenetic tree, all species of Lilium were clustered into two monophyletic groups. Lilium lankongense, L. duchartrei, L. henryi, L. taliense, and Nomocharis pardanthina formed a monophyletic clade, and L. cernuum, L. martagon var. pilosiusculum, L. longiflorum comprised the other monophyletic clade (). This complete cp genome can provide information for population genomic studies, DNA barcoding, and conservation genetics.
Acknowledgements
The authors are really grateful to the opened raw genome data from the public database. The authors thank Dengfeng Xie, Junpei Chen, Haiying Liu, Xin Yang for their assistance in sequencing chloroplast genome and data analysis.
Disclosure statement
The author states no potential conflict of interest and is responsible for the content.
Additional information
Funding
References
- Dierckxsens N, Mardulyn P, Smits G. 2016. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:e18–e18.
- Gao Y-D, Harris A, Zhou S-D, He X-J. 2013. Evolutionary events in Lilium (including Nomocharis, Liliaceae) are temporally correlated with orogenies of the Q-T plateau and the Hengduan Mountains. Mol Phylogenet Evol. 68:443–460.
- Gao Y-D, Harris AJ, He X-J. 2015. Morphological and ecological divergence of Lilium and Nomocharis within the Hengduan Mountains and Qinghai-Tibetan Plateau may result from habitat specialization and hybridization. BMC Evol Biol. 15:147.
- Gao Y-D, Hohenegger M, Harris A, Zhou S-D, He X-J, Wan J. 2012. A new species in the genus Nomocharis Franchet (Liliaceae): evidence that brings the genus Nomocharis into Lilium. Plant Syst Evol. 298:69–85.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Liang SY, Tamura M. 2000. Lilium. In: Wu ZY, Raven PH, editors. Flora of China. Vol. 24. St. Louis: Science Press/Beijing: Missouri Botanical Garden Press; p. 135–149.
- Nishikawa T, Okazaki K, Uchino T, Arakawa K, Nagamine T. 1999. A molecular phylogeny of Lilium in the internal transcribed spacer region of nuclear ribosomal DNA. J Mol Evol. 49:238–249.
- North C, Wills A. 1969. Inter-specific hybrids of Lilium lankongense Franchet produced by embryo-culture. Euphytica. 18:430–434.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.