Abstract
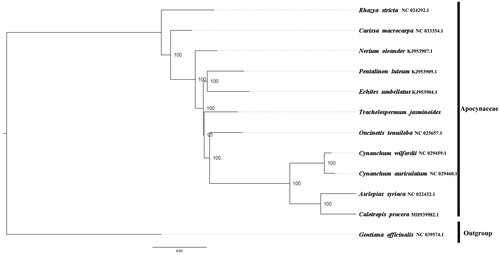
Trachelospermum jasminoides belongs to Apocynaceae. Herein, we reported the complete chloroplast genome of T. jasminoides. To our knowledge, this is the first reported complete chloroplast genome of T. jasminoides. The complete chloroplast genome of T. jasminoides is 155,484 bp in length with a typical quadripartite structure, consisting of a large single-copy region (LSC, 85,514 bp), a single-copy region (SSC, 18,236 bp), and a pair of inverted repeats (IRs, 25,876 bp). There are 114 genes annotated, including 79 unique protein-coding genes, 4 unique ribosomal RNA genes, and 30 transfer RNA genes. To investigate the evolution status of T. jasminoides, as well as Apocynaceae, we constructed a phylogenetic tree with T. jasminoides and other 11 species based on their complete chloroplast genomes. According to the phylogenetic topologies, T. jasminoides was closely related to Oncinotis tenuiloba.
Trachelospermum jasminoides (Lindley) Lemaire is a plant of the family Apocynaceae. It is a lianas woody and distributs in the provinces south of the Yangtze River, such as Anhui, Fujian, Guangdong, Guangxi, Guizhou, etc. (Li et al. Citation1995). Consequently, the genetic and genomic information is urgently needed to promote its systematics research and the development of conservation value of T. jasminoides. As chloroplast carry maternal genes, it plays an important role in phylogeny reconstruction. However, there are no researches about T. jasminoides chloroplast. Here, we report and characterize the complete plastid genome sequence of T. jasminoides (GenBank accession number: MK783315) in an effort to provide genomic resources useful for promoting its conservation.
In this study, fresh leaves of T. jasminoides was collected from Sanlingshan of Zhanjiang city Nature Reserve in Guangdong province of China (110.40°E, 21.18°N). Voucher specimens (Cheng, CL1) were deposited in the Herbarium of Lingnan Normal University, Zhangjiang, China.
The experiment procedure was as reported in Zhu et al. (Citation2018). Around 6 Gb clean data were assembled against the plastome of Trachelospermum asiaticum (MG963252.1) (Fishbein et al. Citation2018) using MITO bim V1.8 (Natural History Museum, University of Oslo, Oslo, Norway) (Hahn et al. Citation2013). The plastome was annotated using Geneious R8.0.2 (Biomatters Ltd., Auckland, New Zealand) against the plastome of T. asiaticum (MG963252.1). The annotation was corrected with DOGMA (Wyman et al. Citation2004).
The plastome of T. jasminoides was found to possess a total length 155,484 bp with the typical quadripartite structure of angiosperms, containing two inverted repeats (IRs) of 25,867 bp, a large single-copy (LSC) region of 85,514 bp and a small single-copy (SSC) region of 18,236 bp. The plastome contains 114 genes, consisting of 79 unique protein-coding genes, 30 unique tRNA genes, and 4 unique rRNA genes. The overall A/T content in the plastome of T. jasminoides is 61.60%, which the corresponding value of the LSC, SSC, and IR region were 63.40, 67.40, and 56.60%, respectively.
We used RAxML (Stamatakis Citation2006) with 1000 bootstraps under the GTRGAMMAI substitution model to reconstruct a maximum likelihood (ML) phylogeny of 10 published complete plastomes of Rubiaceae, using Gentiana officinalis (Gentianaceae) as an outgroup. According to the phylogenetic topologies, T. jasminoides was closely related to Oncinotis tenuiloba. Most nodes in the plastome ML trees were strongly supported (). The complete plastome sequence of T. jasminoides will provide a useful resource for the conservation gengtics of this species as well as for the phylogenetic studies for Apocynaceae.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Li PT, Leeuwenberg AJM, Middleton DJ 1995. Flora of China. Vol. 16. Beijing, Science Press; p. 143–188.
- Fishbein M, Livshultz T, Straub SCK, et al. 2018. Evolution on the backbone: Apocynaceae phylogenomics and new perspectives on growth forms, flowers, and fruits. Am. J. Bot. In press.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads-a baiting and iterative mapping approach. Nucleic Acids Res. 41:e129.
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 22:2688–2690.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
- Zhu ZX, Mu WX, Wang JH, Zhang JR, Zhao KK, Friedman CR, Wang HF. 2018. Complete plastome sequence of Dracaena cambodiana (Asparagaceae): a species considered “Vulnerable” in Southeast Asia. Mitochondr DNA B. 3:620–621.